SNAI2 Antibody - #AF4002
| Product: | SNAI2 Antibody |
| Catalog: | AF4002 |
| Description: | Rabbit polyclonal antibody to SNAI2 |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 30 kDa; 30kD(Calculated). |
| Uniprot: | O43623 |
| RRID: | AB_2835331 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF4002, RRID:AB_2835331.
Fold/Unfold
dJ710H13.1; MGC10182; Neural crest transcription factor Slug; Protein sna; Protein snail homolog 1; Protein snail homolog 2; Protein snail homolog; Slug homolog zinc finger protein; Slug zinc finger protein; SLUGH; SLUGH 1; SLUGH1; SLUGH2; SNA; Sna protein; SNAH; SNAI 2; snai1; SNAI1_HUMAN; Snai2; SNAI2_HUMAN; Snail 2; Snail homolog 1 (Drosophila); Snail homolog 2; Snail2; WS 2D; WS2D; Zinc finger protein SLUG; Zinc finger protein SNAI1; Zinc finger protein SNAI2;
Immunogens
A synthesized peptide derived from human SNAI2, corresponding to a region within the internal amino acids.
Expressed in most adult human tissues, including spleen, thymus, prostate, testis, ovary, small intestine, colon, heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas. Not detected in peripheral blood leukocyte. Expressed in the dermis and in all layers of the epidermis, with high levels of expression in the basal layers (at protein level). Expressed in osteoblasts (at protein level). Expressed in mesenchymal stem cells (at protein level). Expressed in breast tumor cells (at protein level).
- O43623 SNAI2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPRSFLVKKHFNASKKPNYSELDTHTVIISPYLYESYSMPVIPQPEILSSGAYSPITVWTTAAPFHAQLPNGLSPLSGYSSSLGRVSPPPPSDTSSKDHSGSESPISDEEERLQSKLSDPHAIEAEKFQCNLCNKTYSTFSGLAKHKQLHCDAQSRKSFSCKYCDKEYVSLGALKMHIRTHTLPCVCKICGKAFSRPWLLQGHIRTHTGEKPFSCPHCNRAFADRSNLRAHLQTHSDVKKYQCKNCSKTFSRMSLLHKHEESGCCVAH
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional repressor that modulates both activator-dependent and basal transcription. Involved in the generation and migration of neural crest cells. Plays a role in mediating RAF1-induced transcriptional repression of the TJ protein, occludin (OCLN) and subsequent oncogenic transformation of epithelial cells (By similarity). Represses BRCA2 expression by binding to its E2-box-containing silencer and recruiting CTBP1 and HDAC1 in breast cells. In epidermal keratinocytes, binds to the E-box in ITGA3 promoter and represses its transcription. Involved in the regulation of ITGB1 and ITGB4 expression and cell adhesion and proliferation in epidermal keratinocytes. Binds to E-box2 domain of BSG and activates its expression during TGFB1-induced epithelial-mesenchymal transition (EMT) in hepatocytes. Represses E-Cadherin/CDH1 transcription via E-box elements. Involved in osteoblast maturation. Binds to RUNX2 and SOC9 promoters and may act as a positive and negative transcription regulator, respectively, in osteoblasts. Binds to CXCL12 promoter via E-box regions in mesenchymal stem cells and osteoblasts. Plays an essential role in TWIST1-induced EMT and its ability to promote invasion and metastasis.
GSK3B-mediated phosphorylation results in cytoplasmic localization and degradation.
Nucleus. Cytoplasm.
Note: Observed in discrete foci in interphase nuclei. These nuclear foci do not overlap with the nucleoli, the SP100 and the HP1 heterochromatin or the coiled body, suggesting SNAI2 is associated with active transcription or active splicing regions.
Expressed in most adult human tissues, including spleen, thymus, prostate, testis, ovary, small intestine, colon, heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas. Not detected in peripheral blood leukocyte. Expressed in the dermis and in all layers of the epidermis, with high levels of expression in the basal layers (at protein level). Expressed in osteoblasts (at protein level). Expressed in mesenchymal stem cells (at protein level). Expressed in breast tumor cells (at protein level).
Repression activity depends on the C-terminal DNA-binding zinc fingers and on the N-terminal repression domain.
Belongs to the snail C2H2-type zinc-finger protein family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
References
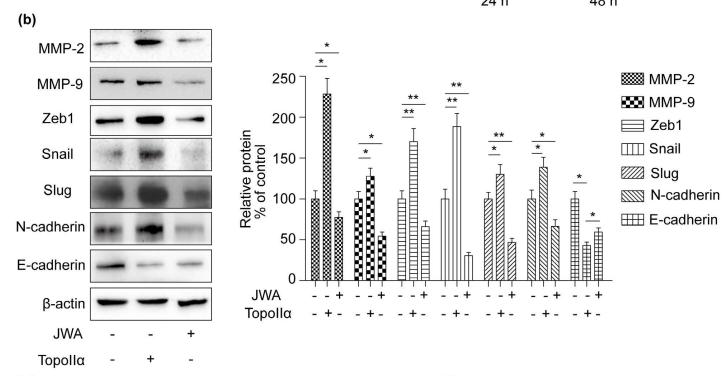
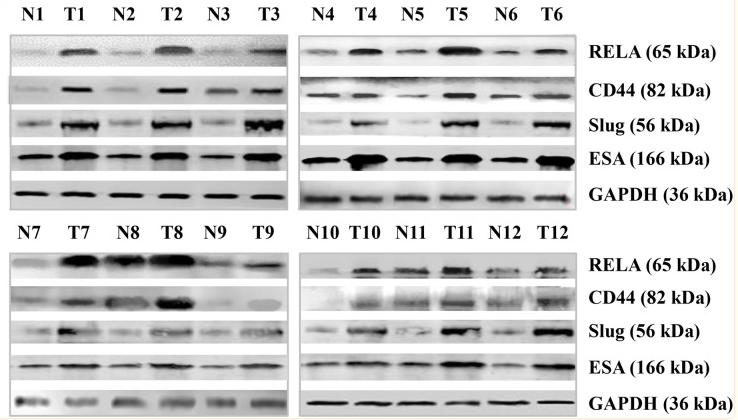
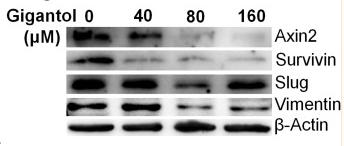
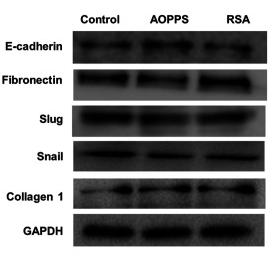
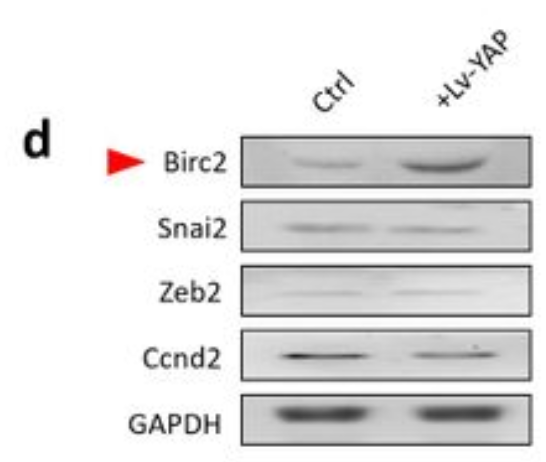
Application: WB Species: Human Sample: breast cancer cells
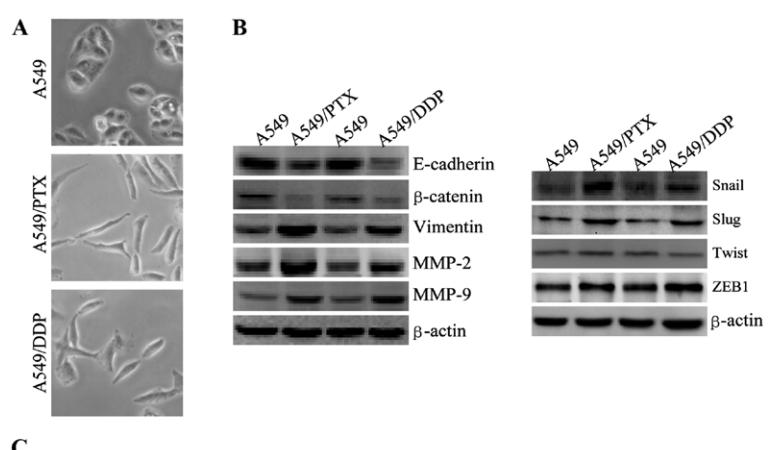
Application: WB Species: human Sample: A549 cells
Application: WB Species: human Sample: A549/PTX cells
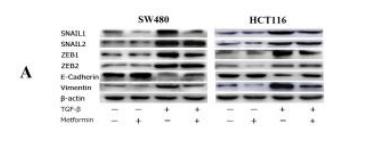
Application: WB Species: human Sample: SW480 and HCT116 cells
Application: WB Species: human Sample: HT29 cells
Application: WB Species: Human Sample: bladder cancer cells
Application: WB Species: Rat Sample: crypt epithelial cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.