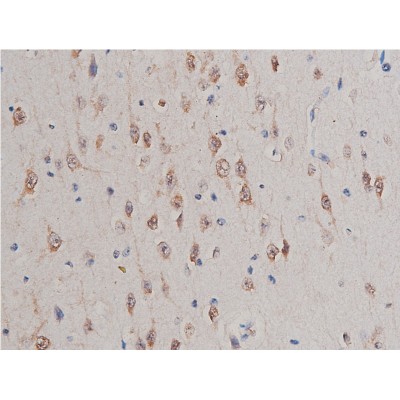
GSK3 alpha/ beta Antibody - #AF6335
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6335, RRID:AB_2835191.
Fold/Unfold
DKFZp686D0638; Glycogen synthase kinase 3 alpha; Glycogen synthase kinase-3 alpha; GSK 3 alpha; GSK 3A; GSK-3 alpha; Gsk3a; GSK3A_HUMAN; GSK3alpha; Serine/threonine protein kinase GSK3A; Glycogen Synthase Kinase 3 Beta; Glycogen synthase kinase-3 beta; GSK 3 beta; GSK-3 beta; GSK3B; GSK3B_HUMAN; GSK3beta isoform; Serine/threonine-protein kinase GSK3B;
Immunogens
A synthesized peptide derived from human GSK3 alpha/ beta, corresponding to a region within the internal amino acids.
Expressed in testis, thymus, prostate and ovary and weakly expressed in lung, brain and kidney. Colocalizes with EIF2AK2/PKR and TAU in the Alzheimer disease (AD) brain.
- P49840 GSK3A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSGGGPSGGGPGGSGRARTSSFAEPGGGGGGGGGGPGGSASGPGGTGGGKASVGAMGGGVGASSSGGGPGGSGGGGSGGPGAGTSFPPPGVKLGRDSGKVTTVVATLGQGPERSQEVAYTDIKVIGNGSFGVVYQARLAETRELVAIKKVLQDKRFKNRELQIMRKLDHCNIVRLRYFFYSSGEKKDELYLNLVLEYVPETVYRVARHFTKAKLTIPILYVKVYMYQLFRSLAYIHSQGVCHRDIKPQNLLVDPDTAVLKLCDFGSAKQLVRGEPNVSYICSRYYRAPELIFGATDYTSSIDVWSAGCVLAELLLGQPIFPGDSGVDQLVEIIKVLGTPTREQIREMNPNYTEFKFPQIKAHPWTKVFKSRTPPEAIALCSSLLEYTPSSRLSPLEACAHSFFDELRCLGTQLPNNRPLPPLFNFSAGELSIQPSLNAILIPPHLRSPAGTTTLTPSSQALTETPTSSDWQSTDATPTLTNSS
- P49841 GSK3B_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSGRPRTTSFAESCKPVQQPSAFGSMKVSRDKDGSKVTTVVATPGQGPDRPQEVSYTDTKVIGNGSFGVVYQAKLCDSGELVAIKKVLQDKRFKNRELQIMRKLDHCNIVRLRYFFYSSGEKKDEVYLNLVLDYVPETVYRVARHYSRAKQTLPVIYVKLYMYQLFRSLAYIHSFGICHRDIKPQNLLLDPDTAVLKLCDFGSAKQLVRGEPNVSYICSRYYRAPELIFGATDYTSSIDVWSAGCVLAELLLGQPIFPGDSGVDQLVEIIKVLGTPTREQIREMNPNYTEFKFPQIKAHPWTKVFRPRTPPEAIALCSRLLEYTPTARLTPLEACAHSFFDELRDPNVKLPNGRDTPALFNFTTQELSSNPPLATILIPPHARIQAAASTPTNATAASDANTGDRGQTNNAASASASNST
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Constitutively active protein kinase that acts as a negative regulator in the hormonal control of glucose homeostasis, Wnt signaling and regulation of transcription factors and microtubules, by phosphorylating and inactivating glycogen synthase (GYS1 or GYS2), CTNNB1/beta-catenin, APC and AXIN1. Requires primed phosphorylation of the majority of its substrates. Contributes to insulin regulation of glycogen synthesis by phosphorylating and inhibiting GYS1 activity and hence glycogen synthesis. Regulates glycogen metabolism in liver, but not in muscle (By similarity). May also mediate the development of insulin resistance by regulating activation of transcription factors. In Wnt signaling, regulates the level and transcriptional activity of nuclear CTNNB1/beta-catenin. Facilitates amyloid precursor protein (APP) processing and the generation of APP-derived amyloid plaques found in Alzheimer disease. May be involved in the regulation of replication in pancreatic beta-cells (By similarity). Is necessary for the establishment of neuronal polarity and axon outgrowth (By similarity). Through phosphorylation of the anti-apoptotic protein MCL1, may control cell apoptosis in response to growth factors deprivation (By similarity). Acts as a regulator of autophagy by mediating phosphorylation of KAT5/TIP60 under starvation conditions, leading to activate KAT5/TIP60 acetyltransferase activity and promote acetylation of key autophagy regulators, such as ULK1 and RUBCNL/Pacer. Negatively regulates extrinsic apoptotic signaling pathway via death domain receptors. Promotes the formation of an anti-apoptotic complex, made of DDX3X, BRIC2 and GSK3B, at death receptors, including TNFRSF10B. The anti-apoptotic function is most effective with weak apoptotic signals and can be overcome by stronger stimulation (By similarity).
Phosphorylated by AKT1 at Ser-21: upon insulin-mediated signaling, the activated PKB/AKT1 protein kinase phosphorylates and desactivates GSK3A, resulting in the dephosphorylation and activation of GYS1. Activated by phosphorylation at Tyr-279.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. GSK-3 subfamily.
Constitutively active protein kinase that acts as a negative regulator in the hormonal control of glucose homeostasis, Wnt signaling and regulation of transcription factors and microtubules, by phosphorylating and inactivating glycogen synthase (GYS1 or GYS2), EIF2B, CTNNB1/beta-catenin, APC, AXIN1, DPYSL2/CRMP2, JUN, NFATC1/NFATC, MAPT/TAU and MACF1. Requires primed phosphorylation of the majority of its substrates. In skeletal muscle, contributes to insulin regulation of glycogen synthesis by phosphorylating and inhibiting GYS1 activity and hence glycogen synthesis. May also mediate the development of insulin resistance by regulating activation of transcription factors. Regulates protein synthesis by controlling the activity of initiation factor 2B (EIF2BE/EIF2B5) in the same manner as glycogen synthase. In Wnt signaling, GSK3B forms a multimeric complex with APC, AXIN1 and CTNNB1/beta-catenin and phosphorylates the N-terminus of CTNNB1 leading to its degradation mediated by ubiquitin/proteasomes. Phosphorylates JUN at sites proximal to its DNA-binding domain, thereby reducing its affinity for DNA. Phosphorylates NFATC1/NFATC on conserved serine residues promoting NFATC1/NFATC nuclear export, shutting off NFATC1/NFATC gene regulation, and thereby opposing the action of calcineurin. Phosphorylates MAPT/TAU on 'Thr-548', decreasing significantly MAPT/TAU ability to bind and stabilize microtubules. MAPT/TAU is the principal component of neurofibrillary tangles in Alzheimer disease. Plays an important role in ERBB2-dependent stabilization of microtubules at the cell cortex. Phosphorylates MACF1, inhibiting its binding to microtubules which is critical for its role in bulge stem cell migration and skin wound repair. Probably regulates NF-kappa-B (NFKB1) at the transcriptional level and is required for the NF-kappa-B-mediated anti-apoptotic response to TNF-alpha (TNF/TNFA). Negatively regulates replication in pancreatic beta-cells, resulting in apoptosis, loss of beta-cells and diabetes. Through phosphorylation of the anti-apoptotic protein MCL1, may control cell apoptosis in response to growth factors deprivation. Phosphorylates MUC1 in breast cancer cells, decreasing the interaction of MUC1 with CTNNB1/beta-catenin. Is necessary for the establishment of neuronal polarity and axon outgrowth. Phosphorylates MARK2, leading to inhibit its activity. Phosphorylates SIK1 at 'Thr-182', leading to sustain its activity. Phosphorylates ZC3HAV1 which enhances its antiviral activity. Phosphorylates SNAI1, leading to its BTRC-triggered ubiquitination and proteasomal degradation. Phosphorylates SFPQ at 'Thr-687' upon T-cell activation. Phosphorylates NR1D1 st 'Ser-55' and 'Ser-59' and stabilizes it by protecting it from proteasomal degradation. Regulates the circadian clock via phosphorylation of the major clock components including ARNTL/BMAL1, CLOCK and PER2. Phosphorylates CLOCK AT 'Ser-427' and targets it for proteasomal degradation. Phosphorylates ARNTL/BMAL1 at 'Ser-17' and 'Ser-21' and primes it for ubiquitination and proteasomal degradation. Phosphorylates OGT at 'Ser-3' or 'Ser-4' which positively regulates its activity. Phosphorylates MYCN in neuroblastoma cells which may promote its degradation. Regulates the circadian rhythmicity of hippocampal long-term potentiation and ARNTL/BMLA1 and PER2 expression (By similarity). Acts as a regulator of autophagy by mediating phosphorylation of KAT5/TIP60 under starvation conditions, leading to activate KAT5/TIP60 acetyltransferase activity and promote acetylation of key autophagy regulators, such as ULK1 and RUBCNL/Pacer. Negatively regulates extrinsic apoptotic signaling pathway via death domain receptors. Promotes the formation of an anti-apoptotic complex, made of DDX3X, BRIC2 and GSK3B, at death receptors, including TNFRSF10B. The anti-apoptotic function is most effective with weak apoptotic signals and can be overcome by stronger stimulation.
Phosphorylated by AKT1 and ILK1. Upon insulin-mediated signaling, the activated PKB/AKT1 protein kinase phosphorylates and desactivates GSK3B, resulting in the dephosphorylation and activation of GYS1. Activated by phosphorylation at Tyr-216. Inactivated by phosphorylation at Ser-9 (Probable). Phosphorylated in a circadian manner in the hippocampus (By similarity).
Mono-ADP-ribosylation by PARP10 negatively regulates kinase activity.
Cytoplasm. Nucleus. Cell membrane.
Note: The phosphorylated form shows localization to cytoplasm and cell membrane. The MEMO1-RHOA-DIAPH1 signaling pathway controls localization of the phosphorylated form to the cell membrane.
Expressed in testis, thymus, prostate and ovary and weakly expressed in lung, brain and kidney. Colocalizes with EIF2AK2/PKR and TAU in the Alzheimer disease (AD) brain.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. GSK-3 subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hedgehog signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Neurodegenerative diseases > Alzheimer's disease.
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Prostate cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Basal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Nervous system > Dopaminergic synapse.
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Melanogenesis.
· Organismal Systems > Endocrine system > Prolactin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.