C/EBP alpha Antibody - #AF6333
| Product: | C/EBP alpha Antibody |
| Catalog: | AF6333 |
| Description: | Rabbit polyclonal antibody to C/EBP alpha |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine |
| Mol.Wt.: | 30,43kDa; 38kD(Calculated). |
| Uniprot: | P49715 |
| RRID: | AB_2835189 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6333, RRID:AB_2835189.
Fold/Unfold
Apoptotic cysteine protease; Apoptotic protease Mch 5; C/EBP alpha; C/ebpalpha; CAP4; Caspase 8 precursor; CBF-A; CCAAT Enhancer Binding Protein alpha; CCAAT/enhancer binding protein (C/EBP), alpha; CCAAT/enhancer-binding protein alpha; CEBP; CEBP A; CEBP alpha; Cebpa; CEBPA_HUMAN; FADD homologous ICE/CED 3 like protease; FADD like ICE; FLICE; ICE like apoptotic protease 5; ICE8; MACH; MCH5; MORT1 associated CED 3 homolog;
Immunogens
A synthesized peptide derived from human C/EBP alpha, corresponding to a region within N-terminal amino acids.
- P49715 CEBPA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MESADFYEAEPRPPMSSHLQSPPHAPSSAAFGFPRGAGPAQPPAPPAAPEPLGGICEHETSIDISAYIDPAAFNDEFLADLFQHSRQQEKAKAAVGPTGGGGGGDFDYPGAPAGPGGAVMPGGAHGPPPGYGCAAAGYLDGRLEPLYERVGAPALRPLVIKQEPREEDEAKQLALAGLFPYQPPPPPPPSHPHPHPPPAHLAAPHLQFQIAHCGQTTMHLQPGHPTPPPTPVPSPHPAPALGAAGLPGPGSALKGLGAAHPDLRASGGSGAGKAKKSVDKNSNEYRVRRERNNIAVRKSRDKAKQRNVETQQKVLELTSDNDRLRKRVEQLSRELDTLRGIFRQLPESSLVKAMGNCA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcription factor that coordinates proliferation arrest and the differentiation of myeloid progenitors, adipocytes, hepatocytes, and cells of the lung and the placenta. Binds directly to the consensus DNA sequence 5'-T[TG]NNGNAA[TG]-3' acting as an activator on distinct target genes. During early embryogenesis, plays essential and redundant functions with CEBPB. Essential for the transition from common myeloid progenitors (CMP) to granulocyte/monocyte progenitors (GMP). Critical for the proper development of the liver and the lung (By similarity). Necessary for terminal adipocyte differentiation, is required for postnatal maintenance of systemic energy homeostasis and lipid storage (By similarity). To regulate these different processes at the proper moment and tissue, interplays with other transcription factors and modulators. Downregulates the expression of genes that maintain cells in an undifferentiated and proliferative state through E2F1 repression, which is critical for its ability to induce adipocyte and granulocyte terminal differentiation. Reciprocally E2F1 blocks adipocyte differentiation by binding to specific promoters and repressing CEBPA binding to its target gene promoters. Proliferation arrest also depends on a functional binding to SWI/SNF complex. In liver, regulates gluconeogenesis and lipogenesis through different mechanisms. To regulate gluconeogenesis, functionally cooperates with FOXO1 binding to IRE-controlled promoters and regulating the expression of target genes such as PCK1 or G6PC. To modulate lipogenesis, interacts and transcriptionally synergizes with SREBF1 in promoter activation of specific lipogenic target genes such as ACAS2. In adipose tissue, seems to act as FOXO1 coactivator accessing to ADIPOQ promoter through FOXO1 binding sites (By similarity).
Can act as dominant-negative. Binds DNA and have transctivation activity, even if much less efficiently than isoform 2. Does not inhibit cell proliferation.
Directly and specifically enhances ribosomal DNA transcription interacting with RNA polymerase I-specific cofactors and inducing histone acetylation.
Phosphorylation at Ser-190 is required for interaction with CDK2, CDK4 and SWI/SNF complex leading to cell cycle inhibition. Dephosphorylated at Ser-190 by protein phosphatase 2A (PP2A) through PI3K/AKT signaling pathway regulation. Phosphorylation at Thr-226 and Thr-230 by GSK3 is constitutive in adipose tissue and lung. In liver, both Thr-226 and Thr-230 are phosphorylated only during feeding but not during fasting. Phosphorylation of the GSK3 consensus sites selectively decreases transactivation activity on IRE-controlled promoters.
Sumoylated, sumoylation blocks the inhibitory effect on cell proliferation by disrupting the interaction with SMARCA2.
Ubiquitinated by COP1 upon interaction with TRIB1.
Nucleus.
Nucleus>Nucleolus.
The recognition sequence (54-72) is required for interaction with TRIB1.
Belongs to the bZIP family. C/EBP subfamily.
Research Fields
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
References
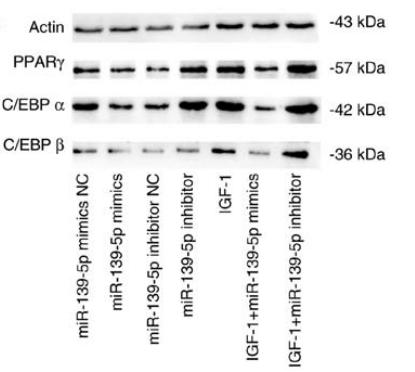
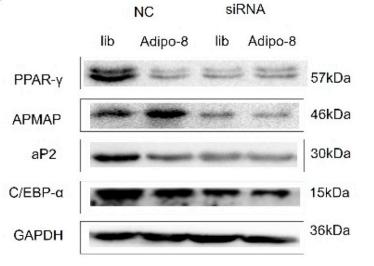
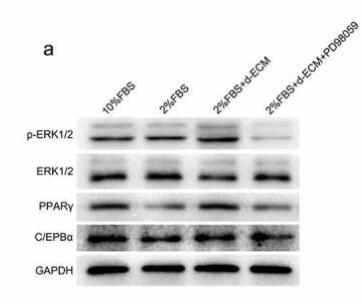
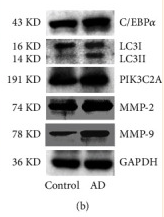
Application: WB Species: Mouse Sample: 3T3-L1 cells
Application: WB Species: human Sample: HemSCs
Application: WB Species: mouse Sample: adipocytes
Application: WB Species: Rat Sample: BMSC
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.