PLK1 Antibody - #AF2685
| Product: | PLK1 Antibody |
| Catalog: | AF2685 |
| Description: | Rabbit polyclonal antibody to PLK1 |
| Application: | WB IHC |
| Cited expt.: | IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 62kDa; 68kD(Calculated). |
| Uniprot: | P53350 |
| RRID: | AB_2847781 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF2685, RRID:AB_2847781.
Fold/Unfold
Cell cycle regulated protein kinase; PLK 1; PLK; PLK-1; plk1; PLK1_HUMAN; Polo like kinase 1; Polo-like kinase 1; Serine/threonine protein kinase 13; Serine/threonine protein kinase PLK1; Serine/threonine-protein kinase 13; Serine/threonine-protein kinase PLK1; STPK 13; STPK13;
Immunogens
A synthesized peptide derived from human PLK1.
- P53350 PLK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSAAVTAGKLARAPADPGKAGVPGVAAPGAPAAAPPAKEIPEVLVDPRSRRRYVRGRFLGKGGFAKCFEISDADTKEVFAGKIVPKSLLLKPHQREKMSMEISIHRSLAHQHVVGFHGFFEDNDFVFVVLELCRRRSLLELHKRRKALTEPEARYYLRQIVLGCQYLHRNRVIHRDLKLGNLFLNEDLEVKIGDFGLATKVEYDGERKKTLCGTPNYIAPEVLSKKGHSFEVDVWSIGCIMYTLLVGKPPFETSCLKETYLRIKKNEYSIPKHINPVAASLIQKMLQTDPTARPTINELLNDEFFTSGYIPARLPITCLTIPPRFSIAPSSLDPSNRKPLTVLNKGLENPLPERPREKEEPVVRETGEVVDCHLSDMLQQLHSVNASKPSERGLVRQEEAEDPACIPIFWVSKWVDYSDKYGLGYQLCDNSVGVLFNDSTRLILYNDGDSLQYIERDGTESYLTVSSHPNSLMKKITLLKYFRNYMSEHLLKAGANITPREGDELARLPYLRTWFRTRSAIILHLSNGSVQINFFQDHTKLILCPLMAAVTYIDEKRDFRTYRLSLLEEYGCCKELASRLRYARTMVDKLLSSRSASNRLKAS
Research Backgrounds
Serine/threonine-protein kinase that performs several important functions throughout M phase of the cell cycle, including the regulation of centrosome maturation and spindle assembly, the removal of cohesins from chromosome arms, the inactivation of anaphase-promoting complex/cyclosome (APC/C) inhibitors, and the regulation of mitotic exit and cytokinesis. Polo-like kinase proteins acts by binding and phosphorylating proteins are that already phosphorylated on a specific motif recognized by the POLO box domains. Phosphorylates BORA, BUB1B/BUBR1, CCNB1, CDC25C, CEP55, ECT2, ERCC6L, FBXO5/EMI1, FOXM1, KIF20A/MKLP2, CENPU, NEDD1, NINL, NPM1, NUDC, PKMYT1/MYT1, KIZ, PPP1R12A/MYPT1, PRC1, RACGAP1/CYK4, SGO1, STAG2/SA2, TEX14, TOPORS, p73/TP73, TPT1, WEE1 and HNRNPU. Plays a key role in centrosome functions and the assembly of bipolar spindles by phosphorylating KIZ, NEDD1 and NINL. NEDD1 phosphorylation promotes subsequent targeting of the gamma-tubulin ring complex (gTuRC) to the centrosome, an important step for spindle formation. Phosphorylation of NINL component of the centrosome leads to NINL dissociation from other centrosomal proteins. Involved in mitosis exit and cytokinesis by phosphorylating CEP55, ECT2, KIF20A/MKLP2, CENPU, PRC1 and RACGAP1. Recruited at the central spindle by phosphorylating and docking PRC1 and KIF20A/MKLP2; creates its own docking sites on PRC1 and KIF20A/MKLP2 by mediating phosphorylation of sites subsequently recognized by the POLO box domains. Phosphorylates RACGAP1, thereby creating a docking site for the Rho GTP exchange factor ECT2 that is essential for the cleavage furrow formation. Promotes the central spindle recruitment of ECT2. Plays a central role in G2/M transition of mitotic cell cycle by phosphorylating CCNB1, CDC25C, FOXM1, CENPU, PKMYT1/MYT1, PPP1R12A/MYPT1 and WEE1. Part of a regulatory circuit that promotes the activation of CDK1 by phosphorylating the positive regulator CDC25C and inhibiting the negative regulators WEE1 and PKMYT1/MYT1. Also acts by mediating phosphorylation of cyclin-B1 (CCNB1) on centrosomes in prophase. Phosphorylates FOXM1, a key mitotic transcription regulator, leading to enhance FOXM1 transcriptional activity. Involved in kinetochore functions and sister chromatid cohesion by phosphorylating BUB1B/BUBR1, FBXO5/EMI1 and STAG2/SA2. PLK1 is high on non-attached kinetochores suggesting a role of PLK1 in kinetochore attachment or in spindle assembly checkpoint (SAC) regulation. Required for kinetochore localization of BUB1B. Regulates the dissociation of cohesin from chromosomes by phosphorylating cohesin subunits such as STAG2/SA2. Phosphorylates SGO1: required for spindle pole localization of isoform 3 of SGO1 and plays a role in regulating its centriole cohesion function. Mediates phosphorylation of FBXO5/EMI1, a negative regulator of the APC/C complex during prophase, leading to FBXO5/EMI1 ubiquitination and degradation by the proteasome. Acts as a negative regulator of p53 family members: phosphorylates TOPORS, leading to inhibit the sumoylation of p53/TP53 and simultaneously enhance the ubiquitination and subsequent degradation of p53/TP53. Phosphorylates the transactivation domain of the transcription factor p73/TP73, leading to inhibit p73/TP73-mediated transcriptional activation and pro-apoptotic functions. Phosphorylates BORA, and thereby promotes the degradation of BORA. Contributes to the regulation of AURKA function. Also required for recovery after DNA damage checkpoint and entry into mitosis. Phosphorylates MISP, leading to stabilization of cortical and astral microtubule attachments required for proper spindle positioning. Together with MEIKIN, acts as a regulator of kinetochore function during meiosis I: required both for mono-orientation of kinetochores on sister chromosomes and protection of centromeric cohesin from separase-mediated cleavage (By similarity). Phosphorylates CEP68 and is required for its degradation. Regulates nuclear envelope breakdown during prophase by phosphorylating DCTN1 resulting in its localization in the nuclear envelope. Phosphorylates the heat shock transcription factor HSF1, promoting HSF1 nuclear translocation upon heat shock. Phosphorylates HSF1 also in the early mitotic period; this phosphorylation regulates HSF1 localization to the spindle pole, the recruitment of the SCF(BTRC) ubiquitin ligase complex induicing HSF1 degradation, and hence mitotic progression. Regulates mitotic progression by phosphorylating RIOK2.
Catalytic activity is enhanced by phosphorylation of Thr-210. Phosphorylation at Thr-210 is first detected on centrosomes in the G2 phase of the cell cycle, peaks in prometaphase and gradually disappears from centrosomes during anaphase. Dephosphorylation at Thr-210 at centrosomes is probably mediated by protein phosphatase 1C (PP1C), via interaction with PPP1R12A/MYPT1. Autophosphorylation and phosphorylation of Ser-137 may not be significant for the activation of PLK1 during mitosis, but may enhance catalytic activity during recovery after DNA damage checkpoint. Phosphorylated in vitro by STK10.
Ubiquitinated by the anaphase promoting complex/cyclosome (APC/C) in anaphase and following DNA damage, leading to its degradation by the proteasome. Ubiquitination is mediated via its interaction with FZR1/CDH1. Ubiquitination and subsequent degradation prevents entry into mitosis and is essential to maintain an efficient G2 DNA damage checkpoint. Monoubiquitination at Lys-492 by the BCR(KLHL22) ubiquitin ligase complex does not lead to degradation: it promotes PLK1 dissociation from phosphoreceptor proteins and subsequent removal from kinetochores, allowing silencing of the spindle assembly checkpoint (SAC) and chromosome segregation.
Nucleus. Chromosome>Centromere>Kinetochore. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome. Cytoplasm>Cytoskeleton>Spindle. Midbody.
Note: localization at the centrosome starts at the G1/S transition (PubMed:24018379). During early stages of mitosis, the phosphorylated form is detected on centrosomes and kinetochores. Localizes to the outer kinetochore. Presence of SGO1 and interaction with the phosphorylated form of BUB1 is required for the kinetochore localization. Localizes onto the central spindle by phosphorylating and docking at midzone proteins KIF20A/MKLP2 and PRC1. Colocalizes with FRY to separating centrosomes and spindle poles from prophase to metaphase in mitosis, but not in other stages of the cell cycle. Localization to the centrosome is required for S phase progression (PubMed:24018379). Colocalizes with HSF1 at the spindle poles during prometaphase (PubMed:18794143).
Placenta and colon.
The POLO box domains act as phosphopeptide-binding module that recognize and bind serine-[phosphothreonine/phosphoserine]-(proline/X) motifs. PLK1 recognizes and binds docking proteins that are already phosphorylated on these motifs, and then phosphorylates them. PLK1 can also create its own docking sites by mediating phosphorylation of serine-[phosphothreonine/phosphoserine]-(proline/X) motifs subsequently recognized by the POLO box domains.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. CDC5/Polo subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
References
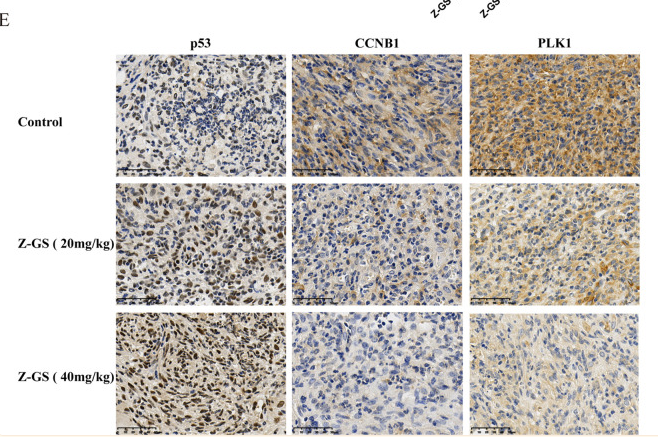
Application: IHC Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.