c-Met Antibody - #AF6128
| Product: | c-Met Antibody |
| Catalog: | AF6128 |
| Description: | Rabbit polyclonal antibody to c-Met |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 155kDa; 156kD(Calculated). |
| Uniprot: | P08581 |
| RRID: | AB_2835011 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6128, RRID:AB_2835011.
Fold/Unfold
AUTS9; c met; D249; Hepatocyte growth factor receptor; HGF; HGF receptor; HGF/SF receptor; HGFR; MET; Met proto oncogene tyrosine kinase; MET proto oncogene, receptor tyrosine kinase; Met proto-oncogene (hepatocyte growth factor receptor); Met proto-oncogene; Met protooncogene; MET_HUMAN; Oncogene MET; Par4; Proto-oncogene c-Met; RCCP2; Scatter factor receptor; SF receptor; Tyrosine-protein kinase Met;
Immunogens
A synthesized peptide derived from human c-Met, corresponding to a region within the internal amino acids.
Expressed in normal hepatocytes as well as in epithelial cells lining the stomach, the small and the large intestine. Found also in basal keratinocytes of esophagus and skin. High levels are found in liver, gastrointestinal tract, thyroid and kidney. Also present in the brain. Expressed in metaphyseal bone (at protein level) (PubMed:26637977).
- P08581 MET_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MKAPAVLAPGILVLLFTLVQRSNGECKEALAKSEMNVNMKYQLPNFTAETPIQNVILHEHHIFLGATNYIYVLNEEDLQKVAEYKTGPVLEHPDCFPCQDCSSKANLSGGVWKDNINMALVVDTYYDDQLISCGSVNRGTCQRHVFPHNHTADIQSEVHCIFSPQIEEPSQCPDCVVSALGAKVLSSVKDRFINFFVGNTINSSYFPDHPLHSISVRRLKETKDGFMFLTDQSYIDVLPEFRDSYPIKYVHAFESNNFIYFLTVQRETLDAQTFHTRIIRFCSINSGLHSYMEMPLECILTEKRKKRSTKKEVFNILQAAYVSKPGAQLARQIGASLNDDILFGVFAQSKPDSAEPMDRSAMCAFPIKYVNDFFNKIVNKNNVRCLQHFYGPNHEHCFNRTLLRNSSGCEARRDEYRTEFTTALQRVDLFMGQFSEVLLTSISTFIKGDLTIANLGTSEGRFMQVVVSRSGPSTPHVNFLLDSHPVSPEVIVEHTLNQNGYTLVITGKKITKIPLNGLGCRHFQSCSQCLSAPPFVQCGWCHDKCVRSEECLSGTWTQQICLPAIYKVFPNSAPLEGGTRLTICGWDFGFRRNNKFDLKKTRVLLGNESCTLTLSESTMNTLKCTVGPAMNKHFNMSIIISNGHGTTQYSTFSYVDPVITSISPKYGPMAGGTLLTLTGNYLNSGNSRHISIGGKTCTLKSVSNSILECYTPAQTISTEFAVKLKIDLANRETSIFSYREDPIVYEIHPTKSFISGGSTITGVGKNLNSVSVPRMVINVHEAGRNFTVACQHRSNSEIICCTTPSLQQLNLQLPLKTKAFFMLDGILSKYFDLIYVHNPVFKPFEKPVMISMGNENVLEIKGNDIDPEAVKGEVLKVGNKSCENIHLHSEAVLCTVPNDLLKLNSELNIEWKQAISSTVLGKVIVQPDQNFTGLIAGVVSISTALLLLLGFFLWLKKRKQIKDLGSELVRYDARVHTPHLDRLVSARSVSPTTEMVSNESVDYRATFPEDQFPNSSQNGSCRQVQYPLTDMSPILTSGDSDISSPLLQNTVHIDLSALNPELVQAVQHVVIGPSSLIVHFNEVIGRGHFGCVYHGTLLDNDGKKIHCAVKSLNRITDIGEVSQFLTEGIIMKDFSHPNVLSLLGICLRSEGSPLVVLPYMKHGDLRNFIRNETHNPTVKDLIGFGLQVAKGMKYLASKKFVHRDLAARNCMLDEKFTVKVADFGLARDMYDKEYYSVHNKTGAKLPVKWMALESLQTQKFTTKSDVWSFGVLLWELMTRGAPPYPDVNTFDITVYLLQGRRLLQPEYCPDPLYEVMLKCWHPKAEMRPSFSELVSRISAIFSTFIGEHYVHVNATYVNVKCVAPYPSLLSSEDNADDEVDTRPASFWETS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Receptor tyrosine kinase that transduces signals from the extracellular matrix into the cytoplasm by binding to hepatocyte growth factor/HGF ligand. Regulates many physiological processes including proliferation, scattering, morphogenesis and survival. Ligand binding at the cell surface induces autophosphorylation of MET on its intracellular domain that provides docking sites for downstream signaling molecules. Following activation by ligand, interacts with the PI3-kinase subunit PIK3R1, PLCG1, SRC, GRB2, STAT3 or the adapter GAB1. Recruitment of these downstream effectors by MET leads to the activation of several signaling cascades including the RAS-ERK, PI3 kinase-AKT, or PLCgamma-PKC. The RAS-ERK activation is associated with the morphogenetic effects while PI3K/AKT coordinates prosurvival effects. During embryonic development, MET signaling plays a role in gastrulation, development and migration of muscles and neuronal precursors, angiogenesis and kidney formation. In adults, participates in wound healing as well as organ regeneration and tissue remodeling. Promotes also differentiation and proliferation of hematopoietic cells. May regulate cortical bone osteogenesis (By similarity).
(Microbial infection) Acts as a receptor for Listeria monocytogenes internalin InlB, mediating entry of the pathogen into cells.
Autophosphorylated in response to ligand binding on Tyr-1234 and Tyr-1235 in the kinase domain leading to further phosphorylation of Tyr-1349 and Tyr-1356 in the C-terminal multifunctional docking site. Dephosphorylated by PTPRJ at Tyr-1349 and Tyr-1365. Dephosphorylated by PTPN1 and PTPN2.
Ubiquitinated. Ubiquitination by CBL regulates MET endocytosis, resulting in decreasing plasma membrane receptor abundance, and in endosomal degradation and/or recycling of internalized receptors.
(Microbial infection) Tyrosine phosphorylation is stimulated by L.monocytogenes InlB. Tyrosine phosphorylation is maximal 10-20 minutes after treatment with InlB and disappears by 60 minutes. The phosphorylated residues were not identified.
Membrane>Single-pass type I membrane protein.
Secreted.
Expressed in normal hepatocytes as well as in epithelial cells lining the stomach, the small and the large intestine. Found also in basal keratinocytes of esophagus and skin. High levels are found in liver, gastrointestinal tract, thyroid and kidney. Also present in the brain. Expressed in metaphyseal bone (at protein level).
The kinase domain is involved in SPSB1 binding.
The beta-propeller Sema domain mediates binding to HGF.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signaling molecules and interaction > Cytokine-cytokine receptor interaction. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > EGFR tyrosine kinase inhibitor resistance.
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
· Human Diseases > Infectious diseases: Bacterial > Epithelial cell signaling in Helicobacter pylori infection.
· Human Diseases > Infectious diseases: Parasitic > Malaria.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
References
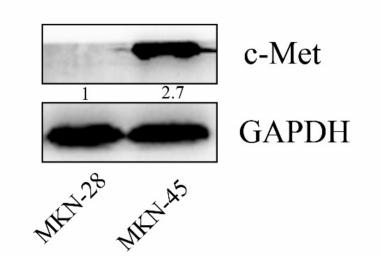
Application: WB Species: human Sample: MKN-45 and MKN-28 cells
Application: WB Species: Human Sample: uterine tissues
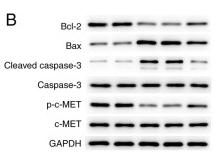
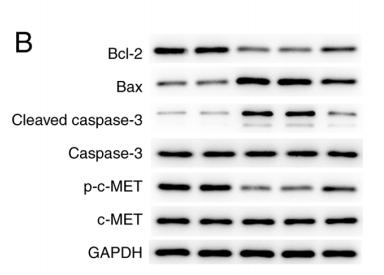
Application: WB Species: Human Sample: gastric cancer cell
Application: WB Species: human Sample: MKN‑45 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.