SP1 Antibody - #AF6121
| Product: | SP1 Antibody |
| Catalog: | AF6121 |
| Description: | Rabbit polyclonal antibody to SP1 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 90kDa; 81kD(Calculated). |
| Uniprot: | P08047 |
| RRID: | AB_2835005 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6121, RRID:AB_2835005.
Fold/Unfold
SP 1; SP1; Sp1 transcription factor; SP1_HUMAN; Specificity protein 1; Transcription factor Sp1; TSFP 1; TSFP1;
Immunogens
A synthesized peptide derived from human SP1, corresponding to a region within the internal amino acids.
Up-regulated in adenocarcinomas of the stomach (at protein level). Isoform 3 is ubiquitously expressed at low levels.
- P08047 SP1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSDQDHSMDEMTAVVKIEKGVGGNNGGNGNGGGAFSQARSSSTGSSSSTGGGGQESQPSPLALLAATCSRIESPNENSNNSQGPSQSGGTGELDLTATQLSQGANGWQIISSSSGATPTSKEQSGSSTNGSNGSESSKNRTVSGGQYVVAAAPNLQNQQVLTGLPGVMPNIQYQVIPQFQTVDGQQLQFAATGAQVQQDGSGQIQIIPGANQQIITNRGSGGNIIAAMPNLLQQAVPLQGLANNVLSGQTQYVTNVPVALNGNITLLPVNSVSAATLTPSSQAVTISSSGSQESGSQPVTSGTTISSASLVSSQASSSSFFTNANSYSTTTTTSNMGIMNFTTSGSSGTNSQGQTPQRVSGLQGSDALNIQQNQTSGGSLQAGQQKEGEQNQQTQQQQILIQPQLVQGGQALQALQAAPLSGQTFTTQAISQETLQNLQLQAVPNSGPIIIRTPTVGPNGQVSWQTLQLQNLQVQNPQAQTITLAPMQGVSLGQTSSSNTTLTPIASAASIPAGTVTVNAAQLSSMPGLQTINLSALGTSGIQVHPIQGLPLAIANAPGDHGAQLGLHGAGGDGIHDDTAGGEEGENSPDAQPQAGRRTRREACTCPYCKDSEGRGSGDPGKKKQHICHIQGCGKVYGKTSHLRAHLRWHTGERPFMCTWSYCGKRFTRSDELQRHKRTHTGEKKFACPECPKRFMRSDHLSKHIKTHQNKKGGPGVALSVGTLPLDSGAGSEGSGTATPSALITTNMVAMEAICPEGIARLANSGINVMQVADLQSINISGNGF
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcription factor that can activate or repress transcription in response to physiological and pathological stimuli. Binds with high affinity to GC-rich motifs and regulates the expression of a large number of genes involved in a variety of processes such as cell growth, apoptosis, differentiation and immune responses. Highly regulated by post-translational modifications (phosphorylations, sumoylation, proteolytic cleavage, glycosylation and acetylation). Binds also the PDGFR-alpha G-box promoter. May have a role in modulating the cellular response to DNA damage. Implicated in chromatin remodeling. Plays an essential role in the regulation of FE65 gene expression. In complex with ATF7IP, maintains telomerase activity in cancer cells by inducing TERT and TERC gene expression. Isoform 3 is a stronger activator of transcription than isoform 1. Positively regulates the transcription of the core clock component ARNTL/BMAL1. Plays a role in the recruitment of SMARCA4/BRG1 on the c-FOS promoter. Plays a role in protecting cells against oxidative stress following brain injury by regulating the expression of RNF112 (By similarity).
Phosphorylated on multiple serine and threonine residues. Phosphorylation is coupled to ubiquitination, sumoylation and proteolytic processing. Phosphorylation on Ser-59 enhances proteolytic cleavage. Phosphorylation on Ser-7 enhances ubiquitination and protein degradation. Hyperphosphorylation on Ser-101 in response to DNA damage has no effect on transcriptional activity. MAPK1/MAPK3-mediated phosphorylation on Thr-453 and Thr-739 enhances VEGF transcription but, represses FGF2-triggered PDGFR-alpha transcription. Also implicated in the repression of RECK by ERBB2. Hyperphosphorylated on Thr-278 and Thr-739 during mitosis by MAPK8 shielding SP1 from degradation by the ubiquitin-dependent pathway. Phosphorylated in the zinc-finger domain by calmodulin-activated PKCzeta. Phosphorylation on Ser-641 by PKCzeta is critical for TSA-activated LHR gene expression through release of its repressor, p107. Phosphorylation on Thr-668, Ser-670 and Thr-681 is stimulated by angiotensin II via the AT1 receptor inducing increased binding to the PDGF-D promoter. This phosphorylation is increased in injured artey wall. Ser-59 and Thr-681 can both be dephosphorylated by PP2A during cell-cycle interphase. Dephosphorylation on Ser-59 leads to increased chromatin association during interphase and increases the transcriptional activity. On insulin stimulation, sequentially glycosylated and phosphorylated on several C-terminal serine and threonine residues.
Acetylated. Acetylation/deacetylation events affect transcriptional activity. Deacetylation leads to an increase in the expression the 12(s)-lipooxygenase gene though recruitment of p300 to the promoter.
Ubiquitinated. Ubiquitination occurs on the C-terminal proteolytically-cleaved peptide and is triggered by phosphorylation.
Sumoylated with SUMO1. Sumoylation modulates proteolytic cleavage of the N-terminal repressor domain. Sumoylation levels are attenuated during tumorigenesis. Phosphorylation mediates SP1 desumoylation.
Proteolytic cleavage in the N-terminal repressor domain is prevented by sumoylation. The C-terminal cleaved product is susceptible to degradation.
O-glycosylated; Contains 8 N-acetylglucosamine side chains. Levels are controlled by insulin and the SP1 phosphorylation states. Insulin-mediated O-glycosylation locates SP1 to the nucleus, where it is sequentially deglycosylated and phosphorylated. O-glycosylation affects transcriptional activity through disrupting the interaction with a number of transcription factors including ELF1 and NFYA. Also inhibits interaction with the HIV1 promoter. Inhibited by peroxisomome proliferator receptor gamma (PPARgamma).
Nucleus. Cytoplasm.
Note: Nuclear location is governed by glycosylated/phosphorylated states. Insulin promotes nuclear location, while glucagon favors cytoplasmic location.
Up-regulated in adenocarcinomas of the stomach (at protein level). Isoform 3 is ubiquitously expressed at low levels.
The 9aaTAD motif is a transactivation domain present in a large number of yeast and animal transcription factors.
Belongs to the Sp1 C2H2-type zinc-finger protein family.
Research Fields
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Neurodegenerative diseases > Huntington's disease.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Overview > Choline metabolism in cancer. (View pathway)
· Organismal Systems > Endocrine system > Estrogen signaling pathway. (View pathway)
References
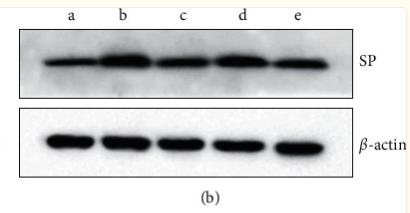
Application: WB Species: Rat Sample: colon
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.