Phospho-MKK3/MKK6 (Ser189/Thr193) Antibody - #AF3805
| Product: | Phospho-MKK3/MKK6 (Ser189/Thr193) Antibody |
| Catalog: | AF3805 |
| Description: | Rabbit polyclonal antibody to Phospho-MKK3/MKK6 (Ser189/Thr193) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 39kD,37kD(Calculated). |
| Uniprot: | P46734 | P52564 |
| RRID: | AB_2847119 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3805, RRID:AB_2847119.
Fold/Unfold
AW212142; dual specificity mitogen activated protein kinase kinase 3; Dual specificity mitogen-activated protein kinase kinase 3; MAP kinase kinase 3; map2k3; MAPK ERK kinase 3; MAPK/ERK kinase 3; MAPKK 3; MAPKK3; MEK 3; MEK3; Mitogen activated protein kinase kinase 3; MKK 3; MKK3; mMKK3b; MP2K3_HUMAN; PRKMK 3; PRKMK3; protein kinase, mitogen-activated, kinase 3; SAPK kinase 2; SAPKK 2; SAPKK2; Stress activated protein kinase kinase 2; Dual specificity mitogen activated protein kinase kinase 6; Dual specificity mitogen-activated protein kinase kinase 6; MAP kinase kinase 6; MAP2K6; MAPK/ERK kinase 6; MAPKK 6; MAPKK6; MEK 6; MEK6; Mitogen Activated Protein Kinase Kinase 6; MKK 6; MKK6; MP2K6_HUMAN; PRKMK6; Protein kinase mitogen activated kinase 6; protein kinase, mitogen-activated, kinase 6 (MAP kinase kinase 6); SAPK kinase 3; SAPKK-3; SAPKK3; SKK3; Stress-activated protein kinase kinase 3;
Immunogens
A synthesized peptide derived from human MKK3/MKK6 around the phosphorylation site of Ser189/Thr193.
Abundant expression is seen in the skeletal muscle. It is also widely expressed in other tissues.
P52564 MP2K6_HUMAN:Isoform 2 is only expressed in skeletal muscle. Isoform 1 is expressed in skeletal muscle, heart, and in lesser extent in liver or pancreas.
- P46734 MP2K3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MESPASSQPASMPQSKGKSKRKKDLRISCMSKPPAPNPTPPRNLDSRTFITIGDRNFEVEADDLVTISELGRGAYGVVEKVRHAQSGTIMAVKRIRATVNSQEQKRLLMDLDINMRTVDCFYTVTFYGALFREGDVWICMELMDTSLDKFYRKVLDKNMTIPEDILGEIAVSIVRALEHLHSKLSVIHRDVKPSNVLINKEGHVKMCDFGISGYLVDSVAKTMDAGCKPYMAPERINPELNQKGYNVKSDVWSLGITMIEMAILRFPYESWGTPFQQLKQVVEEPSPQLPADRFSPEFVDFTAQCLRKNPAERMSYLELMEHPFFTLHKTKKTDIAAFVKEILGEDS
- P52564 MP2K6_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSQSKGKKRNPGLKIPKEAFEQPQTSSTPPRDLDSKACISIGNQNFEVKADDLEPIMELGRGAYGVVEKMRHVPSGQIMAVKRIRATVNSQEQKRLLMDLDISMRTVDCPFTVTFYGALFREGDVWICMELMDTSLDKFYKQVIDKGQTIPEDILGKIAVSIVKALEHLHSKLSVIHRDVKPSNVLINALGQVKMCDFGISGYLVDSVAKTIDAGCKPYMAPERINPELNQKGYSVKSDIWSLGITMIELAILRFPYDSWGTPFQQLKQVVEEPSPQLPADKFSAEFVDFTSQCLKKNSKERPTYPELMQHPFFTLHESKGTDVASFVKLILGD
Research Backgrounds
Dual specificity kinase. Is activated by cytokines and environmental stress in vivo. Catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in the MAP kinase p38. Part of a signaling cascade that begins with the activation of the adrenergic receptor ADRA1B and leads to the activation of MAPK14.
Autophosphorylated. Phosphorylation on Ser-218 and Thr-222 by MAP kinase kinase kinases regulates positively the kinase activity. Phosphorylated by TAOK2.
Yersinia yopJ may acetylate Ser/Thr residues, preventing phosphorylation and activation, thus blocking the MAPK signaling pathway.
Abundant expression is seen in the skeletal muscle. It is also widely expressed in other tissues.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase subfamily.
Dual specificity protein kinase which acts as an essential component of the MAP kinase signal transduction pathway. With MAP3K3/MKK3, catalyzes the concomitant phosphorylation of a threonine and a tyrosine residue in the MAP kinases p38 MAPK11, MAPK12, MAPK13 and MAPK14 and plays an important role in the regulation of cellular responses to cytokines and all kinds of stresses. Especially, MAP2K3/MKK3 and MAP2K6/MKK6 are both essential for the activation of MAPK11 and MAPK13 induced by environmental stress, whereas MAP2K6/MKK6 is the major MAPK11 activator in response to TNF. MAP2K6/MKK6 also phosphorylates and activates PAK6. The p38 MAP kinase signal transduction pathway leads to direct activation of transcription factors. Nuclear targets of p38 MAP kinase include the transcription factors ATF2 and ELK1. Within the p38 MAPK signal transduction pathway, MAP3K6/MKK6 mediates phosphorylation of STAT4 through MAPK14 activation, and is therefore required for STAT4 activation and STAT4-regulated gene expression in response to IL-12 stimulation. The pathway is also crucial for IL-6-induced SOCS3 expression and down-regulation of IL-6-mediated gene induction; and for IFNG-dependent gene transcription. Has a role in osteoclast differentiation through NF-kappa-B transactivation by TNFSF11, and in endochondral ossification and since SOX9 is another likely downstream target of the p38 MAPK pathway. MAP2K6/MKK6 mediates apoptotic cell death in thymocytes. Acts also as a regulator for melanocytes dendricity, through the modulation of Rho family GTPases.
Weakly autophosphorylated. Phosphorylated at Ser-207 and Thr-211 by the majority of M3Ks, such as MAP3K5/ASK1, MAP3K1/MEKK1, MAP3K2/MEKK2, MAP3K3/MEKK3, MAP3K4/MEKK4, MAP3K7/TAK1, MAP3K11/MLK3 and MAP3K17/TAOK2.
Acetylation of Ser-207 and Thr-211 by Yersinia yopJ prevents phosphorylation and activation, thus blocking the MAPK signaling pathway.
Nucleus. Cytoplasm. Cytoplasm>Cytoskeleton.
Note: Binds to microtubules.
Isoform 2 is only expressed in skeletal muscle. Isoform 1 is expressed in skeletal muscle, heart, and in lesser extent in liver or pancreas.
The DVD domain (residues 311-334) contains a conserved docking site and is found in the mammalian MAP kinase kinases (MAP2Ks). The DVD sites bind to their specific upstream MAP kinase kinase kinases (MAP3Ks) and are essential for activation.
The D domain (residues 4-19) contains a conserved docking site and is required for the binding to MAPK substrates.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Neurodegenerative diseases > Amyotrophic lateral sclerosis (ALS).
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Viral > Influenza A.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Sensory system > Inflammatory mediator regulation of TRP channels. (View pathway)
References
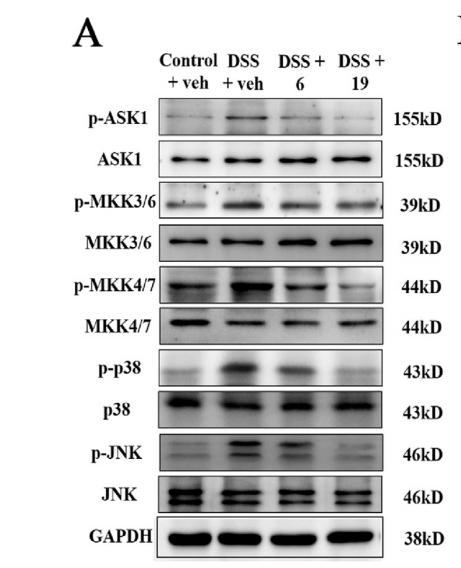
Application: WB Species: mice Sample: colon tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.