Phospho-Parkin (Ser65) Antibody - #AF3500
| Product: | Phospho-Parkin (Ser65) Antibody |
| Catalog: | AF3500 |
| Description: | Rabbit polyclonal antibody to Phospho-Parkin (Ser65) |
| Application: | WB IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 52kD(Calculated). |
| Uniprot: | O60260 |
| RRID: | AB_2846814 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3500, RRID:AB_2846814.
Fold/Unfold
AR JP; E3 ubiquitin ligase; E3 ubiquitin protein ligase parkin; E3 ubiquitin-protein ligase parkin; FRA6E; LPRS 2; LPRS2; PARK 2; Park2; Parkin 2; Parkinson disease (autosomal recessive juvenile) 2; Parkinson disease (autosomal recessive, juvenile) 2, parkin; Parkinson disease protein 2; Parkinson juvenile disease protein 2; Parkinson protein 2 E3 ubiquitin protein ligase; Parkinson protein 2, E3 ubiquitin protein ligase (parkin); PDJ; PRKN 2; PRKN; PRKN2; PRKN2_HUMAN; Ubiquitin E3 ligase PRKN;
Immunogens
A synthesized peptide derived from human Parkin around the phosphorylation site of Ser65.
Highly expressed in the brain including the substantia nigra. Expressed in heart, testis and skeletal muscle. Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients. Overexpression protects dopamine neurons from kainate-mediated apoptosis. Found in serum (at protein level).
- O60260 PRKN_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MIVFVRFNSSHGFPVEVDSDTSIFQLKEVVAKRQGVPADQLRVIFAGKELRNDWTVQNCDLDQQSIVHIVQRPWRKGQEMNATGGDDPRNAAGGCEREPQSLTRVDLSSSVLPGDSVGLAVILHTDSRKDSPPAGSPAGRSIYNSFYVYCKGPCQRVQPGKLRVQCSTCRQATLTLTQGPSCWDDVLIPNRMSGECQSPHCPGTSAEFFFKCGAHPTSDKETSVALHLIATNSRNITCITCTDVRSPVLVFQCNSRHVICLDCFHLYCVTRLNDRQFVHDPQLGYSLPCVAGCPNSLIKELHHFRILGEEQYNRYQQYGAEECVLQMGGVLCPRPGCGAGLLPEPDQRKVTCEGGNGLGCGFAFCRECKEAYHEGECSAVFEASGTTTQAYRVDERAAEQARWEAASKETIKKTTKPCPRCHVPVEKNGGCMHMKCPQPQCRLEWCWNCGCEWNRVCMGDHWFDV
Research Backgrounds
Functions within a multiprotein E3 ubiquitin ligase complex, catalyzing the covalent attachment of ubiquitin moieties onto substrate proteins, such as BCL2, SYT11, CCNE1, GPR37, RHOT1/MIRO1, MFN1, MFN2, STUB1, SNCAIP, SEPTIN5, TOMM20, USP30, ZNF746 and AIMP2. Mediates monoubiquitination as well as 'Lys-6', 'Lys-11', 'Lys-48'-linked and 'Lys-63'-linked polyubiquitination of substrates depending on the context. Participates in the removal and/or detoxification of abnormally folded or damaged protein by mediating 'Lys-63'-linked polyubiquitination of misfolded proteins such as PARK7: 'Lys-63'-linked polyubiquitinated misfolded proteins are then recognized by HDAC6, leading to their recruitment to aggresomes, followed by degradation. Mediates 'Lys-63'-linked polyubiquitination of a 22 kDa O-linked glycosylated isoform of SNCAIP, possibly playing a role in Lewy-body formation. Mediates monoubiquitination of BCL2, thereby acting as a positive regulator of autophagy. Promotes the autophagic degradation of dysfunctional depolarized mitochondria (mitophagy) by promoting the ubiquitination of mitochondrial proteins such as TOMM20, RHOT1/MIRO1 and USP30. Preferentially assembles 'Lys-6'-, 'Lys-11'- and 'Lys-63'-linked polyubiquitin chains following mitochondrial damage, leading to mitophagy. Mediates 'Lys-48'-linked polyubiquitination of ZNF746, followed by degradation of ZNF746 by the proteasome; possibly playing a role in the regulation of neuron death. Limits the production of reactive oxygen species (ROS). Regulates cyclin-E during neuronal apoptosis. In collaboration with CHPF isoform 2, may enhance cell viability and protect cells from oxidative stress. Independently of its ubiquitin ligase activity, protects from apoptosis by the transcriptional repression of p53/TP53. May protect neurons against alpha synuclein toxicity, proteasomal dysfunction, GPR37 accumulation, and kainate-induced excitotoxicity. May play a role in controlling neurotransmitter trafficking at the presynaptic terminal and in calcium-dependent exocytosis. May represent a tumor suppressor gene.
Auto-ubiquitinates in an E2-dependent manner leading to its own degradation. Also polyubiquitinated by RNF41 for proteasomal degradation.
S-nitrosylated. The inhibition of PRKN ubiquitin E3 ligase activity by S-nitrosylation could contribute to the degenerative process in PD by impairing the ubiquitination of PRKN substrates.
Phosphorylation at Ser-65 by PINK1 contributes to activate PRKN activity. It is however not sufficient and requires binding to phosphorylated ubiquitin as well.
Cytoplasm>Cytosol. Nucleus. Endoplasmic reticulum. Mitochondrion.
Note: Mainly localizes in the cytosol. Co-localizes with SYT11 in neutrites. Co-localizes with SNCAIP in brainstem Lewy bodies. Mitochondrial localization gradually increases with cellular growth. Also relocates to dysfunctional mitochondria that have lost the mitochondrial membrane potential; recruitment to mitochondria is PINK1-dependent.
Highly expressed in the brain including the substantia nigra. Expressed in heart, testis and skeletal muscle. Expression is down-regulated or absent in tumor biopsies, and absent in the brain of PARK2 patients. Overexpression protects dopamine neurons from kainate-mediated apoptosis. Found in serum (at protein level).
The ubiquitin-like domain binds the PSMD4 subunit of 26S proteasomes.
The RING-type 1 zinc finger domain is required to repress p53/TP53 transcription.
Members of the RBR family are atypical E3 ligases. They interact with the E2 conjugating enzyme UBE2L3 and function like HECT-type E3 enzymes: they bind E2s via the first RING domain, but require an obligate trans-thiolation step during the ubiquitin transfer, requiring a conserved cysteine residue in the second RING domain (PubMed:21532592).
Belongs to the RBR family. Parkin subfamily.
Research Fields
· Genetic Information Processing > Folding, sorting and degradation > Ubiquitin mediated proteolysis. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > Protein processing in endoplasmic reticulum. (View pathway)
· Human Diseases > Neurodegenerative diseases > Parkinson's disease.
References
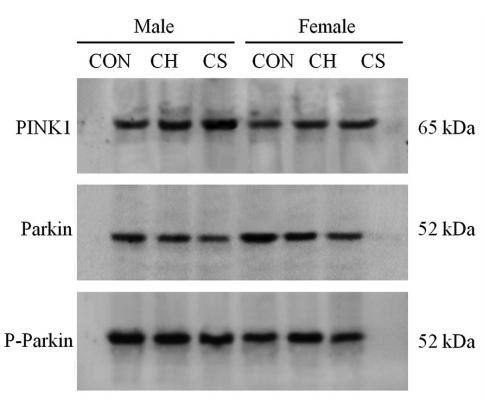
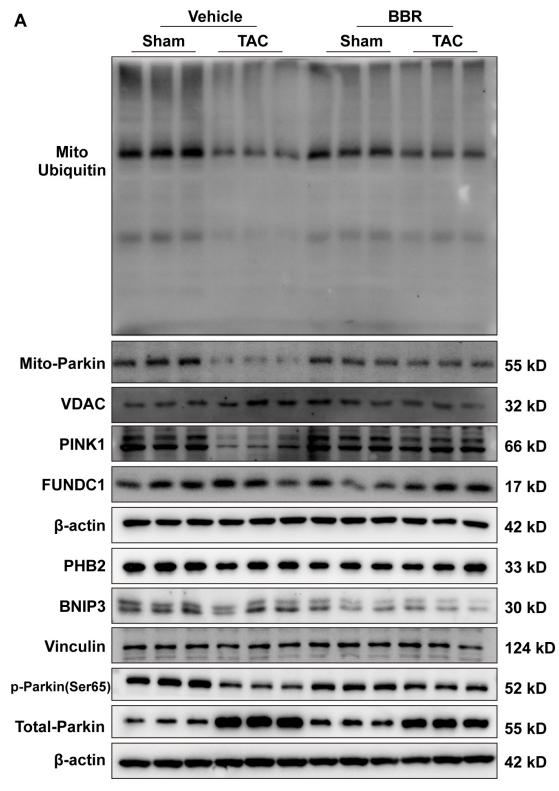
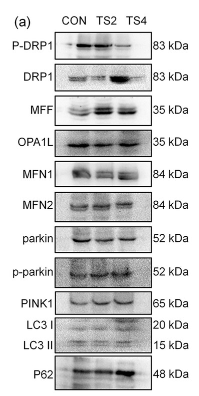
Application: WB Species: mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.