pan Sodium Channel Antibody - #AF0255
| Product: | pan Sodium Channel Antibody |
| Catalog: | AF0255 |
| Description: | Rabbit polyclonal antibody to pan Sodium Channel |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Bovine, Horse, Sheep, Rabbit, Chicken |
| Mol.Wt.: | 230kDa; 229kD,208kD,227kD,226kD,228kD,205kD,225kD,221kD(Calculated). |
| Uniprot: | P35498 | P35499 | Q14524 | Q15858 | Q99250 | Q9NY46 | Q9UI33 | Q9UQD0 | Q9Y5Y9 |
| RRID: | AB_2833430 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0255, RRID:AB_2833430.
Fold/Unfold
HBSC II;HH1;hNaN;hNE-Na;hPN3;KIAA1356;MED;NAC1;NAC2;NAC3;NENA;Neuroendocrine sodium channel;Peripheral nerve sodium channel 3;Peripheral nerve sodium channel 5;Peripheral sodium channel 1;PN1;PN3;PN5;SCN1;SCN10A;SCN11A;SCN12A;SCN1A;SCN2A;SCN2A1;SCN2A2;SCN3A;SCN4A;SCN5A;SCN8A;SCN9A;Sensory neuron sodium channel 2;SkM1;SNS2;Sodium channel protein brain I subunit alpha;Sodium channel protein brain II subunit alpha;Sodium channel protein brain III subunit alpha;Sodium channel protein cardiac muscle subunit alpha;Sodium channel protein skeletal muscle subunit alpha;Sodium channel protein type 1 subunit alpha;Sodium channel protein type 10 subunit alpha;Sodium channel protein type 11 subunit alpha;Sodium channel protein type 2 subunit alpha;Sodium channel protein type 3 subunit alpha;Sodium channel protein type 4 subunit alpha;Sodium channel protein type 5 subunit alpha;Sodium channel protein type 8 subunit alpha;Sodium channel protein type 9 subunit alpha;Sodium channel protein type II subunit alpha;Sodium channel protein type III subunit alpha;Sodium channel protein type IV subunit alpha;Sodium channel protein type IX subunit alpha;Sodium channel protein type V subunit alpha;Sodium channel protein type VIII subunit alpha;Sodium channel protein type X subunit alpha;Sodium channel protein type XI subunit alpha;Voltage-gated sodium channel subtype III;Voltage-gated sodium channel subunit alpha Nav1.1;Voltage-gated sodium channel subunit alpha Nav1.2;Voltage-gated sodium channel subunit alpha Nav1.3;Voltage-gated sodium channel subunit alpha Nav1.4;Voltage-gated sodium channel subunit alpha Nav1.5;Voltage-gated sodium channel subunit alpha Nav1.6;Voltage-gated sodium channel subunit alpha Nav1.7;Voltage-gated sodium channel subunit alpha Nav1.8;Voltage-gated sodium channel subunit alpha Nav1.9;
Immunogens
A synthesized peptide derived from human pan Sodium Channel, corresponding to a region within the internal amino acids.
P35498(SCN1A_HUMAN) >>Visit HPA database.
P35499(SCN4A_HUMAN) >>Visit HPA database.
Q14524(SCN5A_HUMAN) >>Visit HPA database.
Q15858(SCN9A_HUMAN) >>Visit HPA database.
Q99250(SCN2A_HUMAN) >>Visit HPA database.
Q9NY46(SCN3A_HUMAN) >>Visit HPA database.
Q9UI33(SCNBA_HUMAN) >>Visit HPA database.
Found in jejunal circular smooth muscle cells (at protein level). Expressed in human atrial and ventricular cardiac muscle but not in adult skeletal muscle, brain, myometrium, liver, or spleen. Isoform 4 is expressed in brain.
Q15858 SCN9A_HUMAN:Expressed strongly in dorsal root ganglion, with only minor levels elsewhere in the body, smooth muscle cells, MTC cell line and C-cell carcinoma. Isoform 1 is expressed preferentially in the central and peripheral nervous system. Isoform 2 is expressed preferentially in the dorsal root ganglion.
Q9NY46 SCN3A_HUMAN:Expressed in enterochromaffin cells in both colon and small bowel (at protein level).
Q9UI33 SCNBA_HUMAN:Expressed in the dorsal root ganglia and trigeminal ganglia, olfactory bulb, hippocampus, cerebellar cortex, spinal cord, spleen, small intestine and placenta.
Q9UQD0 SCN8A_HUMAN:Expressed in the hippocampus with increased expression in epileptic tissue compared to normal adjacent tissue (at protein level) (PubMed:28842554). Isoform 5: Expressed in non-neuronal tissues, such as monocytes/macrophages.
Q9Y5Y9 SCNAA_HUMAN:Expressed in the dorsal root ganglia and sciatic nerve.
- P35498 SCN1A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEQTVLVPPGPDSFNFFTRESLAAIERRIAEEKAKNPKPDKKDDDENGPKPNSDLEAGKNLPFIYGDIPPEMVSEPLEDLDPYYINKKTFIVLNKGKAIFRFSATSALYILTPFNPLRKIAIKILVHSLFSMLIMCTILTNCVFMTMSNPPDWTKNVEYTFTGIYTFESLIKIIARGFCLEDFTFLRDPWNWLDFTVITFAYVTEFVDLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCIQWPPTNASLEEHSIEKNITVNYNGTLINETVFEFDWKSYIQDSRYHYFLEGFLDALLCGNSSDAGQCPEGYMCVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDFWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLINLILAVVAMAYEEQNQATLEEAEQKEAEFQQMIEQLKKQQEAAQQAATATASEHSREPSAAGRLSDSSSEASKLSSKSAKERRNRRKKRKQKEQSGGEEKDEDEFQKSESEDSIRRKGFRFSIEGNRLTYEKRYSSPHQSLLSIRGSLFSPRRNSRTSLFSFRGRAKDVGSENDFADDEHSTFEDNESRRDSLFVPRRHGERRNSNLSQTSRSSRMLAVFPANGKMHSTVDCNGVVSLVGGPSVPTSPVGQLLPEVIIDKPATDDNGTTTETEMRKRRSSSFHVSMDFLEDPSQRQRAMSIASILTNTVEELEESRQKCPPCWYKFSNIFLIWDCSPYWLKVKHVVNLVVMDPFVDLAITICIVLNTLFMAMEHYPMTDHFNNVLTVGNLVFTGIFTAEMFLKIIAMDPYYYFQEGWNIFDGFIVTLSLVELGLANVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKDCVCKIASDCQLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLTVFMMVMVIGNLVVLNLFLALLLSSFSADNLAATDDDNEMNNLQIAVDRMHKGVAYVKRKIYEFIQQSFIRKQKILDEIKPLDDLNNKKDSCMSNHTAEIGKDLDYLKDVNGTTSGIGTGSSVEKYIIDESDYMSFINNPSLTVTVPIAVGESDFENLNTEDFSSESDLEESKEKLNESSSSSEGSTVDIGAPVEEQPVVEPEETLEPEACFTEGCVQRFKCCQINVEEGRGKQWWNLRRTCFRIVEHNWFETFIVFMILLSSGALAFEDIYIDQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGYQTYFTNAWCWLDFLIVDVSLVSLTANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCINTTTGDRFDIEDVNNHTDCLKLIERNETARWKNVKVNFDNVGFGYLSLLQVATFKGWMDIMYAAVDSRNVELQPKYEESLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPGNKFQGMVFDFVTRQVFDISIMILICLNMVTMMVETDDQSEYVTTILSRINLVFIVLFTGECVLKLISLRHYYFTIGWNIFDFVVVILSIVGMFLAELIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKREVGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSKPPDCDPNKVNPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFMEFEKLSQFAAALEPPLNLPQPNKLQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEERFMASNPSKVSYQPITTTLKRKQEEVSAVIIQRAYRRHLLKRTVKQASFTYNKNKIKGGANLLIKEDMIIDRINENSITEKTDLTMSTAACPPSYDRVTKPIVEKHEQEGKDEKAKGK
- P35499 SCN4A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MARPSLCTLVPLGPECLRPFTRESLAAIEQRAVEEEARLQRNKQMEIEEPERKPRSDLEAGKNLPMIYGDPPPEVIGIPLEDLDPYYSNKKTFIVLNKGKAIFRFSATPALYLLSPFSVVRRGAIKVLIHALFSMFIMITILTNCVFMTMSDPPPWSKNVEYTFTGIYTFESLIKILARGFCVDDFTFLRDPWNWLDFSVIMMAYLTEFVDLGNISALRTFRVLRALKTITVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALVGLQLFMGNLRQKCVRWPPPFNDTNTTWYSNDTWYGNDTWYGNEMWYGNDSWYANDTWNSHASWATNDTFDWDAYISDEGNFYFLEGSNDALLCGNSSDAGHCPEGYECIKTGRNPNYGYTSYDTFSWAFLALFRLMTQDYWENLFQLTLRAAGKTYMIFFVVIIFLGSFYLINLILAVVAMAYAEQNEATLAEDKEKEEEFQQMLEKFKKHQEELEKAKAAQALEGGEADGDPAHGKDCNGSLDTSQGEKGAPRQSSSGDSGISDAMEELEEAHQKCPPWWYKCAHKVLIWNCCAPWLKFKNIIHLIVMDPFVDLGITICIVLNTLFMAMEHYPMTEHFDNVLTVGNLVFTGIFTAEMVLKLIAMDPYEYFQQGWNIFDSIIVTLSLVELGLANVQGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKIALDCNLPRWHMHDFFHSFLIVFRILCGEWIETMWDCMEVAGQAMCLTVFLMVMVIGNLVVLNLFLALLLSSFSADSLAASDEDGEMNNLQIAIGRIKLGIGFAKAFLLGLLHGKILSPKDIMLSLGEADGAGEAGEAGETAPEDEKKEPPEEDLKKDNHILNHMGLADGPPSSLELDHLNFINNPYLTIQVPIASEESDLEMPTEEETDTFSEPEDSKKPPQPLYDGNSSVCSTADYKPPEEDPEEQAEENPEGEQPEECFTEACVQRWPCLYVDISQGRGKKWWTLRRACFKIVEHNWFETFIVFMILLSSGALAFEDIYIEQRRVIRTILEYADKVFTYIFIMEMLLKWVAYGFKVYFTNAWCWLDFLIVDVSIISLVANWLGYSELGPIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYYCINTTTSERFDISEVNNKSECESLMHTGQVRWLNVKVNYDNVGLGYLSLLQVATFKGWMDIMYAAVDSREKEEQPQYEVNLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGKDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPQNKIQGMVYDLVTKQAFDITIMILICLNMVTMMVETDNQSQLKVDILYNINMIFIIIFTGECVLKMLALRQYYFTVGWNIFDFVVVILSIVGLALSDLIQKYFVSPTLFRVIRLARIGRVLRLIRGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMSNFAYVKKESGIDDMFNFETFGNSIICLFEITTSAGWDGLLNPILNSGPPDCDPNLENPGTSVKGDCGNPSIGICFFCSYIIISFLIVVNMYIAIILENFNVATEESSEPLGEDDFEMFYETWEKFDPDATQFIAYSRLSDFVDTLQEPLRIAKPNKIKLITLDLPMVPGDKIHCLDILFALTKEVLGDSGEMDALKQTMEEKFMAANPSKVSYEPITTTLKRKHEEVCAIKIQRAYRRHLLQRSMKQASYMYRHSHDGSGDDAPEKEGLLANTMSKMYGHENGNSSSPSPEEKGEAGDAGPTMGLMPISPSDTAWPPAPPPGQTVRPGVKESLV
- Q14524 SCN5A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MANFLLPRGTSSFRRFTRESLAAIEKRMAEKQARGSTTLQESREGLPEEEAPRPQLDLQASKKLPDLYGNPPQELIGEPLEDLDPFYSTQKTFIVLNKGKTIFRFSATNALYVLSPFHPIRRAAVKILVHSLFNMLIMCTILTNCVFMAQHDPPPWTKYVEYTFTAIYTFESLVKILARGFCLHAFTFLRDPWNWLDFSVIIMAYTTEFVDLGNVSALRTFRVLRALKTISVISGLKTIVGALIQSVKKLADVMVLTVFCLSVFALIGLQLFMGNLRHKCVRNFTALNGTNGSVEADGLVWESLDLYLSDPENYLLKNGTSDVLLCGNSSDAGTCPEGYRCLKAGENPDHGYTSFDSFAWAFLALFRLMTQDCWERLYQQTLRSAGKIYMIFFMLVIFLGSFYLVNLILAVVAMAYEEQNQATIAETEEKEKRFQEAMEMLKKEHEALTIRGVDTVSRSSLEMSPLAPVNSHERRSKRRKRMSSGTEECGEDRLPKSDSEDGPRAMNHLSLTRGLSRTSMKPRSSRGSIFTFRRRDLGSEADFADDENSTAGESESHHTSLLVPWPLRRTSAQGQPSPGTSAPGHALHGKKNSTVDCNGVVSLLGAGDPEATSPGSHLLRPVMLEHPPDTTTPSEEPGGPQMLTSQAPCVDGFEEPGARQRALSAVSVLTSALEELEESRHKCPPCWNRLAQRYLIWECCPLWMSIKQGVKLVVMDPFTDLTITMCIVLNTLFMALEHYNMTSEFEEMLQVGNLVFTGIFTAEMTFKIIALDPYYYFQQGWNIFDSIIVILSLMELGLSRMSNLSVLRSFRLLRVFKLAKSWPTLNTLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKNYSELRDSDSGLLPRWHMMDFFHAFLIIFRILCGEWIETMWDCMEVSGQSLCLLVFLLVMVIGNLVVLNLFLALLLSSFSADNLTAPDEDREMNNLQLALARIQRGLRFVKRTTWDFCCGLLRQRPQKPAALAAQGQLPSCIATPYSPPPPETEKVPPTRKETRFEEGEQPGQGTPGDPEPVCVPIAVAESDTDDQEEDEENSLGTEEESSKQQESQPVSGGPEAPPDSRTWSQVSATASSEAEASASQADWRQQWKAEPQAPGCGETPEDSCSEGSTADMTNTAELLEQIPDLGQDVKDPEDCFTEGCVRRCPCCAVDTTQAPGKVWWRLRKTCYHIVEHSWFETFIIFMILLSSGALAFEDIYLEERKTIKVLLEYADKMFTYVFVLEMLLKWVAYGFKKYFTNAWCWLDFLIVDVSLVSLVANTLGFAEMGPIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFGRCINQTEGDLPLNYTIVNNKSQCESLNLTGELYWTKVKVNFDNVGAGYLALLQVATFKGWMDIMYAAVDSRGYEEQPQWEYNLYMYIYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKYQGFIFDIVTKQAFDVTIMFLICLNMVTMMVETDDQSPEKINILAKINLLFVAIFTGECIVKLAALRHYYFTNSWNIFDFVVVILSIVGTVLSDIIQKYFFSPTLFRVIRLARIGRILRLIRGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMANFAYVKWEAGIDDMFNFQTFANSMLCLFQITTSAGWDGLLSPILNTGPPYCDPTLPNSNGSRGDCGSPAVGILFFTTYIIISFLIVVNMYIAIILENFSVATEESTEPLSEDDFDMFYEIWEKFDPEATQFIEYSVLSDFADALSEPLRIAKPNQISLINMDLPMVSGDRIHCMDILFAFTKRVLGESGEMDALKIQMEEKFMAANPSKISYEPITTTLRRKHEEVSAMVIQRAFRRHLLQRSLKHASFLFRQQAGSGLSEEDAPEREGLIAYVMSENFSRPLGPPSSSSISSTSFPPSYDSVTRATSDNLQVRGSDYSHSEDLADFPPSPDRDRESIV
- Q15858 SCN9A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAMLPPPGPQSFVHFTKQSLALIEQRIAERKSKEPKEEKKDDDEEAPKPSSDLEAGKQLPFIYGDIPPGMVSEPLEDLDPYYADKKTFIVLNKGKTIFRFNATPALYMLSPFSPLRRISIKILVHSLFSMLIMCTILTNCIFMTMNNPPDWTKNVEYTFTGIYTFESLVKILARGFCVGEFTFLRDPWNWLDFVVIVFAYLTEFVNLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLKHKCFRNSLENNETLESIMNTLESEEDFRKYFYYLEGSKDALLCGFSTDSGQCPEGYTCVKIGRNPDYGYTSFDTFSWAFLALFRLMTQDYWENLYQQTLRAAGKTYMIFFVVVIFLGSFYLINLILAVVAMAYEEQNQANIEEAKQKELEFQQMLDRLKKEQEEAEAIAAAAAEYTSIRRSRIMGLSESSSETSKLSSKSAKERRNRRKKKNQKKLSSGEEKGDAEKLSKSESEDSIRRKSFHLGVEGHRRAHEKRLSTPNQSPLSIRGSLFSARRSSRTSLFSFKGRGRDIGSETEFADDEHSIFGDNESRRGSLFVPHRPQERRSSNISQASRSPPMLPVNGKMHSAVDCNGVVSLVDGRSALMLPNGQLLPEVIIDKATSDDSGTTNQIHKKRRCSSYLLSEDMLNDPNLRQRAMSRASILTNTVEELEESRQKCPPWWYRFAHKFLIWNCSPYWIKFKKCIYFIVMDPFVDLAITICIVLNTLFMAMEHHPMTEEFKNVLAIGNLVFTGIFAAEMVLKLIAMDPYEYFQVGWNIFDSLIVTLSLVELFLADVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINDDCTLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLIVYMMVMVIGNLVVLNLFLALLLSSFSSDNLTAIEEDPDANNLQIAVTRIKKGINYVKQTLREFILKAFSKKPKISREIRQAEDLNTKKENYISNHTLAEMSKGHNFLKEKDKISGFGSSVDKHLMEDSDGQSFIHNPSLTVTVPIAPGESDLENMNAEELSSDSDSEYSKVRLNRSSSSECSTVDNPLPGEGEEAEAEPMNSDEPEACFTDGCVWRFSCCQVNIESGKGKIWWNIRKTCYKIVEHSWFESFIVLMILLSSGALAFEDIYIERKKTIKIILEYADKIFTYIFILEMLLKWIAYGYKTYFTNAWCWLDFLIVDVSLVTLVANTLGYSDLGPIKSLRTLRALRPLRALSRFEGMRVVVNALIGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYECINTTDGSRFPASQVPNRSECFALMNVSQNVRWKNLKVNFDNVGLGYLSLLQVATFKGWTIIMYAAVDSVNVDKQPKYEYSLYMYIYFVVFIIFGSFFTLNLFIGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPGNKIQGCIFDLVTNQAFDISIMVLICLNMVTMMVEKEGQSQHMTEVLYWINVVFIILFTGECVLKLISLRHYYFTVGWNIFDFVVVIISIVGMFLADLIETYFVSPTLFRVIRLARIGRILRLVKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKKEDGINDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSKPPDCDPKKVHPGSSVEGDCGNPSVGIFYFVSYIIISFLVVVNMYIAVILENFSVATEESTEPLSEDDFEMFYEVWEKFDPDATQFIEFSKLSDFAAALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDSLRSQMEERFMSANPSKVSYEPITTTLKRKQEDVSATVIQRAYRRYRLRQNVKNISSIYIKDGDRDDDLLNKKDMAFDNVNENSSPEKTDATSSTTSPPSYDSVTKPDKEKYEQDRTEKEDKGKDSKESKK
- Q99250 SCN2A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQSVLVPPGPDSFRFFTRESLAAIEQRIAEEKAKRPKQERKDEDDENGPKPNSDLEAGKSLPFIYGDIPPEMVSVPLEDLDPYYINKKTFIVLNKGKAISRFSATPALYILTPFNPIRKLAIKILVHSLFNMLIMCTILTNCVFMTMSNPPDWTKNVEYTFTGIYTFESLIKILARGFCLEDFTFLRDPWNWLDFTVITFAYVTEFVDLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCLQWPPDNSSFEINITSFFNNSLDGNGTTFNRTVSIFNWDEYIEDKSHFYFLEGQNDALLCGNSSDAGQCPEGYICVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDFWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLINLILAVVAMAYEEQNQATLEEAEQKEAEFQQMLEQLKKQQEEAQAAAAAASAESRDFSGAGGIGVFSESSSVASKLSSKSEKELKNRRKKKKQKEQSGEEEKNDRVRKSESEDSIRRKGFRFSLEGSRLTYEKRFSSPHQSLLSIRGSLFSPRRNSRASLFSFRGRAKDIGSENDFADDEHSTFEDNDSRRDSLFVPHRHGERRHSNVSQASRASRVLPILPMNGKMHSAVDCNGVVSLVGGPSTLTSAGQLLPEGTTTETEIRKRRSSSYHVSMDLLEDPTSRQRAMSIASILTNTMEELEESRQKCPPCWYKFANMCLIWDCCKPWLKVKHLVNLVVMDPFVDLAITICIVLNTLFMAMEHYPMTEQFSSVLSVGNLVFTGIFTAEMFLKIIAMDPYYYFQEGWNIFDGFIVSLSLMELGLANVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKISNDCELPRWHMHDFFHSFLIVFRVLCGEWIETMWDCMEVAGQTMCLTVFMMVMVIGNLVVLNLFLALLLSSFSSDNLAATDDDNEMNNLQIAVGRMQKGIDFVKRKIREFIQKAFVRKQKALDEIKPLEDLNNKKDSCISNHTTIEIGKDLNYLKDGNGTTSGIGSSVEKYVVDESDYMSFINNPSLTVTVPIAVGESDFENLNTEEFSSESDMEESKEKLNATSSSEGSTVDIGAPAEGEQPEVEPEESLEPEACFTEDCVRKFKCCQISIEEGKGKLWWNLRKTCYKIVEHNWFETFIVFMILLSSGALAFEDIYIEQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGFQVYFTNAWCWLDFLIVDVSLVSLTANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALLGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCINYTTGEMFDVSVVNNYSECKALIESNQTARWKNVKVNFDNVGLGYLSLLQVATFKGWMDIMYAAVDSRNVELQPKYEDNLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPANKFQGMVFDFVTKQVFDISIMILICLNMVTMMVETDDQSQEMTNILYWINLVFIVLFTGECVLKLISLRYYYFTIGWNIFDFVVVILSIVGMFLAELIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKREVGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSGPPDCDPDKDHPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFIEFAKLSDFADALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEERFMASNPSKVSYEPITTTLKRKQEEVSAIIIQRAYRRYLLKQKVKKVSSIYKKDKGKECDGTPIKEDTLIDKLNENSTPEKTDMTPSTTSPPSYDSVTKPEKEKFEKDKSEKEDKGKDIRESKK
- Q9NY46 SCN3A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAQALLVPPGPESFRLFTRESLAAIEKRAAEEKAKKPKKEQDNDDENKPKPNSDLEAGKNLPFIYGDIPPEMVSEPLEDLDPYYINKKTFIVMNKGKAIFRFSATSALYILTPLNPVRKIAIKILVHSLFSMLIMCTILTNCVFMTLSNPPDWTKNVEYTFTGIYTFESLIKILARGFCLEDFTFLRDPWNWLDFSVIVMAYVTEFVSLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCLQWPPSDSAFETNTTSYFNGTMDSNGTFVNVTMSTFNWKDYIGDDSHFYVLDGQKDPLLCGNGSDAGQCPEGYICVKAGRNPNYGYTSFDTFSWAFLSLFRLMTQDYWENLYQLTLRAAGKTYMIFFVLVIFLGSFYLVNLILAVVAMAYEEQNQATLEEAEQKEAEFQQMLEQLKKQQEEAQAVAAASAASRDFSGIGGLGELLESSSEASKLSSKSAKEWRNRRKKRRQREHLEGNNKGERDSFPKSESEDSVKRSSFLFSMDGNRLTSDKKFCSPHQSLLSIRGSLFSPRRNSKTSIFSFRGRAKDVGSENDFADDEHSTFEDSESRRDSLFVPHRHGERRNSNVSQASMSSRMVPGLPANGKMHSTVDCNGVVSLVGGPSALTSPTGQLPPEGTTTETEVRKRRLSSYQISMEMLEDSSGRQRAVSIASILTNTMEELEESRQKCPPCWYRFANVFLIWDCCDAWLKVKHLVNLIVMDPFVDLAITICIVLNTLFMAMEHYPMTEQFSSVLTVGNLVFTGIFTAEMVLKIIAMDPYYYFQEGWNIFDGIIVSLSLMELGLSNVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINDDCTLPRWHMNDFFHSFLIVFRVLCGEWIETMWDCMEVAGQTMCLIVFMLVMVIGNLVVLNLFLALLLSSFSSDNLAATDDDNEMNNLQIAVGRMQKGIDYVKNKMRECFQKAFFRKPKVIEIHEGNKIDSCMSNNTGIEISKELNYLRDGNGTTSGVGTGSSVEKYVIDENDYMSFINNPSLTVTVPIAVGESDFENLNTEEFSSESELEESKEKLNATSSSEGSTVDVVLPREGEQAETEPEEDLKPEACFTEGCIKKFPFCQVSTEEGKGKIWWNLRKTCYSIVEHNWFETFIVFMILLSSGALAFEDIYIEQRKTIKTMLEYADKVFTYIFILEMLLKWVAYGFQTYFTNAWCWLDFLIVDVSLVSLVANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFYHCVNMTTGNMFDISDVNNLSDCQALGKQARWKNVKVNFDNVGAGYLALLQVATFKGWMDIMYAAVDSRDVKLQPVYEENLYMYLYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPANKFQGMVFDFVTRQVFDISIMILICLNMVTMMVETDDQGKYMTLVLSRINLVFIVLFTGEFVLKLVSLRHYYFTIGWNIFDFVVVILSIVGMFLAEMIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYAIFGMSNFAYVKKEAGIDDMFNFETFGNSMICLFQITTSAGWDGLLAPILNSAPPDCDPDTIHPGSSVKGDCGNPSVGIFFFVSYIIISFLVVVNMYIAVILENFSVATEESAEPLSEDDFEMFYEVWEKFDPDATQFIEFSKLSDFAAALDPPLLIAKPNKVQLIAMDLPMVSGDRIHCLDILFAFTKRVLGESGEMDALRIQMEDRFMASNPSKVSYEPITTTLKRKQEEVSAAIIQRNFRCYLLKQRLKNISSNYNKEAIKGRIDLPIKQDMIIDKLNGNSTPEKTDGSSSTTSPPSYDSVTKPDKEKFEKDKPEKESKGKEVRENQK
- Q9UI33 SCNBA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDDRCYPVIFPDERNFRPFTSDSLAAIEKRIAIQKEKKKSKDQTGEVPQPRPQLDLKASRKLPKLYGDIPRELIGKPLEDLDPFYRNHKTFMVLNRKRTIYRFSAKHALFIFGPFNSIRSLAIRVSVHSLFSMFIIGTVIINCVFMATGPAKNSNSNNTDIAECVFTGIYIFEALIKILARGFILDEFSFLRDPWNWLDSIVIGIAIVSYIPGITIKLLPLRTFRVFRALKAISVVSRLKVIVGALLRSVKKLVNVIILTFFCLSIFALVGQQLFMGSLNLKCISRDCKNISNPEAYDHCFEKKENSPEFKMCGIWMGNSACSIQYECKHTKINPDYNYTNFDNFGWSFLAMFRLMTQDSWEKLYQQTLRTTGLYSVFFFIVVIFLGSFYLINLTLAVVTMAYEEQNKNVAAEIEAKEKMFQEAQQLLKEEKEALVAMGIDRSSLTSLETSYFTPKKRKLFGNKKRKSFFLRESGKDQPPGSDSDEDCQKKPQLLEQTKRLSQNLSLDHFDEHGDPLQRQRALSAVSILTITMKEQEKSQEPCLPCGENLASKYLVWNCCPQWLCVKKVLRTVMTDPFTELAITICIIINTVFLAMEHHKMEASFEKMLNIGNLVFTSIFIAEMCLKIIALDPYHYFRRGWNIFDSIVALLSFADVMNCVLQKRSWPFLRSFRVLRVFKLAKSWPTLNTLIKIIGNSVGALGSLTVVLVIVIFIFSVVGMQLFGRSFNSQKSPKLCNPTGPTVSCLRHWHMGDFWHSFLVVFRILCGEWIENMWECMQEANASSSLCVIVFILITVIGKLVVLNLFIALLLNSFSNEERNGNLEGEARKTKVQLALDRFRRAFCFVRHTLEHFCHKWCRKQNLPQQKEVAGGCAAQSKDIIPLVMEMKRGSETQEELGILTSVPKTLGVRHDWTWLAPLAEEEDDVEFSGEDNAQRITQPEPEQQAYELHQENKKPTSQRVQSVEIDMFSEDEPHLTIQDPRKKSDVTSILSECSTIDLQDGFGWLPEMVPKKQPERCLPKGFGCCFPCCSVDKRKPPWVIWWNLRKTCYQIVKHSWFESFIIFVILLSSGALIFEDVHLENQPKIQELLNCTDIIFTHIFILEMVLKWVAFGFGKYFTSAWCCLDFIIVIVSVTTLINLMELKSFRTLRALRPLRALSQFEGMKVVVNALIGAIPAILNVLLVCLIFWLVFCILGVYFFSGKFGKCINGTDSVINYTIITNKSQCESGNFSWINQKVNFDNVGNAYLALLQVATFKGWMDIIYAAVDSTEKEQQPEFESNSLGYIYFVVFIIFGSFFTLNLFIGVIIDNFNQQQKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKCQGLVFDIVTSQIFDIIIISLIILNMISMMAESYNQPKAMKSILDHLNWVFVVIFTLECLIKIFALRQYYFTNGWNLFDCVVVLLSIVSTMISTLENQEHIPFPPTLFRIVRLARIGRILRLVRAARGIRTLLFALMMSLPSLFNIGLLLFLIMFIYAILGMNWFSKVNPESGIDDIFNFKTFASSMLCLFQISTSAGWDSLLSPMLRSKESCNSSSENCHLPGIATSYFVSYIIISFLIVVNMYIAVILENFNTATEESEDPLGEDDFDIFYEVWEKFDPEATQFIKYSALSDFADALPEPLRVAKPNKYQFLVMDLPMVSEDRLHCMDILFAFTARVLGGSDGLDSMKAMMEEKFMEANPLKKLYEPIVTTTKRKEEERGAAIIQKAFRKYMMKVTKGDQGDQNDLENGPHSPLQTLCNGDLSSFGVAKGKVHCD
- Q9UQD0 SCN8A_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAARLLAPPGPDSFKPFTPESLANIERRIAESKLKKPPKADGSHREDDEDSKPKPNSDLEAGKSLPFIYGDIPQGLVAVPLEDFDPYYLTQKTFVVLNRGKTLFRFSATPALYILSPFNLIRRIAIKILIHSVFSMIIMCTILTNCVFMTFSNPPDWSKNVEYTFTGIYTFESLVKIIARGFCIDGFTFLRDPWNWLDFSVIMMAYITEFVNLGNVSALRTFRVLRALKTISVIPGLKTIVGALIQSVKKLSDVMILTVFCLSVFALIGLQLFMGNLRNKCVVWPINFNESYLENGTKGFDWEEYINNKTNFYTVPGMLEPLLCGNSSDAGQCPEGYQCMKAGRNPNYGYTSFDTFSWAFLALFRLMTQDYWENLYQLTLRAAGKTYMIFFVLVIFVGSFYLVNLILAVVAMAYEEQNQATLEEAEQKEAEFKAMLEQLKKQQEEAQAAAMATSAGTVSEDAIEEEGEEGGGSPRSSSEISKLSSKSAKERRNRRKKRKQKELSEGEEKGDPEKVFKSESEDGMRRKAFRLPDNRIGRKFSIMNQSLLSIPGSPFLSRHNSKSSIFSFRGPGRFRDPGSENEFADDEHSTVEESEGRRDSLFIPIRARERRSSYSGYSGYSQGSRSSRIFPSLRRSVKRNSTVDCNGVVSLIGGPGSHIGGRLLPEATTEVEIKKKGPGSLLVSMDQLASYGRKDRINSIMSVVTNTLVEELEESQRKCPPCWYKFANTFLIWECHPYWIKLKEIVNLIVMDPFVDLAITICIVLNTLFMAMEHHPMTPQFEHVLAVGNLVFTGIFTAEMFLKLIAMDPYYYFQEGWNIFDGFIVSLSLMELSLADVEGLSVLRSFRLLRVFKLAKSWPTLNMLIKIIGNSVGALGNLTLVLAIIVFIFAVVGMQLFGKSYKECVCKINQDCELPRWHMHDFFHSFLIVFRVLCGEWIETMWDCMEVAGQAMCLIVFMMVMVIGNLVVLNLFLALLLSSFSADNLAATDDDGEMNNLQISVIRIKKGVAWTKLKVHAFMQAHFKQREADEVKPLDELYEKKANCIANHTGADIHRNGDFQKNGNGTTSGIGSSVEKYIIDEDHMSFINNPNLTVRVPIAVGESDFENLNTEDVSSESDPEGSKDKLDDTSSSEGSTIDIKPEVEEVPVEQPEEYLDPDACFTEGCVQRFKCCQVNIEEGLGKSWWILRKTCFLIVEHNWFETFIIFMILLSSGALAFEDIYIEQRKTIRTILEYADKVFTYIFILEMLLKWTAYGFVKFFTNAWCWLDFLIVAVSLVSLIANALGYSELGAIKSLRTLRALRPLRALSRFEGMRVVVNALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKYHYCFNETSEIRFEIEDVNNKTECEKLMEGNNTEIRWKNVKINFDNVGAGYLALLQVATFKGWMDIMYAAVDSRKPDEQPKYEDNIYMYIYFVIFIIFGSFFTLNLFIGVIIDNFNQQKKKFGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKIQGIVFDFVTQQAFDIVIMMLICLNMVTMMVETDTQSKQMENILYWINLVFVIFFTCECVLKMFALRHYYFTIGWNIFDFVVVILSIVGMFLADIIEKYFVSPTLFRVIRLARIGRILRLIKGAKGIRTLLFALMMSLPALFNIGLLLFLVMFIFSIFGMSNFAYVKHEAGIDDMFNFETFGNSMICLFQITTSAGWDGLLLPILNRPPDCSLDKEHPGSGFKGDCGNPSVGIFFFVSYIIISFLIVVNMYIAIILENFSVATEESADPLSEDDFETFYEIWEKFDPDATQFIEYCKLADFADALEHPLRVPKPNTIELIAMDLPMVSGDRIHCLDILFAFTKRVLGDSGELDILRQQMEERFVASNPSKVSYEPITTTLRRKQEEVSAVVLQRAYRGHLARRGFICKKTTSNKLENGGTHREKKESTPSTASLPSYDSVTKPEKEKQQRAEEGRRERAKRQKEVRESKC
- Q9Y5Y9 SCNAA_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEFPIGSLETNNFRRFTPESLVEIEKQIAAKQGTKKAREKHREQKDQEEKPRPQLDLKACNQLPKFYGELPAELIGEPLEDLDPFYSTHRTFMVLNKGRTISRFSATRALWLFSPFNLIRRTAIKVSVHSWFSLFITVTILVNCVCMTRTDLPEKIEYVFTVIYTFEALIKILARGFCLNEFTYLRDPWNWLDFSVITLAYVGTAIDLRGISGLRTFRVLRALKTVSVIPGLKVIVGALIHSVKKLADVTILTIFCLSVFALVGLQLFKGNLKNKCVKNDMAVNETTNYSSHRKPDIYINKRGTSDPLLCGNGSDSGHCPDGYICLKTSDNPDFNYTSFDSFAWAFLSLFRLMTQDSWERLYQQTLRTSGKIYMIFFVLVIFLGSFYLVNLILAVVTMAYEEQNQATTDEIEAKEKKFQEALEMLRKEQEVLAALGIDTTSLHSHNGSPLTSKNASERRHRIKPRVSEGSTEDNKSPRSDPYNQRRMSFLGLASGKRRASHGSVFHFRSPGRDISLPEGVTDDGVFPGDHESHRGSLLLGGGAGQQGPLPRSPLPQPSNPDSRHGEDEHQPPPTSELAPGAVDVSAFDAGQKKTFLSAEYLDEPFRAQRAMSVVSIITSVLEELEESEQKCPPCLTSLSQKYLIWDCCPMWVKLKTILFGLVTDPFAELTITLCIVVNTIFMAMEHHGMSPTFEAMLQIGNIVFTIFFTAEMVFKIIAFDPYYYFQKKWNIFDCIIVTVSLLELGVAKKGSLSVLRSFRLLRVFKLAKSWPTLNTLIKIIGNSVGALGNLTIILAIIVFVFALVGKQLLGENYRNNRKNISAPHEDWPRWHMHDFFHSFLIVFRILCGEWIENMWACMEVGQKSICLILFLTVMVLGNLVVLNLFIALLLNSFSADNLTAPEDDGEVNNLQVALARIQVFGHRTKQALCSFFSRSCPFPQPKAEPELVVKLPLSSSKAENHIAANTARGSSGGLQAPRGPRDEHSDFIANPTVWVSVPIAEGESDLDDLEDDGGEDAQSFQQEVIPKGQQEQLQQVERCGDHLTPRSPGTGTSSEDLAPSLGETWKDESVPQVPAEGVDDTSSSEGSTVDCLDPEEILRKIPELADDLEEPDDCFTEGCIRHCPCCKLDTTKSPWDVGWQVRKTCYRIVEHSWFESFIIFMILLSSGSLAFEDYYLDQKPTVKALLEYTDRVFTFIFVFEMLLKWVAYGFKKYFTNAWCWLDFLIVNISLISLTAKILEYSEVAPIKALRTLRALRPLRALSRFEGMRVVVDALVGAIPSIMNVLLVCLIFWLIFSIMGVNLFAGKFWRCINYTDGEFSLVPLSIVNNKSDCKIQNSTGSFFWVNVKVNFDNVAMGYLALLQVATFKGWMDIMYAAVDSREVNMQPKWEDNVYMYLYFVIFIIFGGFFTLNLFVGVIIDNFNQQKKKLGGQDIFMTEEQKKYYNAMKKLGSKKPQKPIPRPLNKFQGFVFDIVTRQAFDITIMVLICLNMITMMVETDDQSEEKTKILGKINQFFVAVFTGECVMKMFALRQYYFTNGWNVFDFIVVVLSIASLIFSAILKSLQSYFSPTLFRVIRLARIGRILRLIRAAKGIRTLLFALMMSLPALFNIGLLLFLVMFIYSIFGMSSFPHVRWEAGIDDMFNFQTFANSMLCLFQITTSAGWDGLLSPILNTGPPYCDPNLPNSNGTRGDCGSPAVGIIFFTTYIIISFLIMVNMYIAVILENFNVATEESTEPLSEDDFDMFYETWEKFDPEATQFITFSALSDFADTLSGPLRIPKPNRNILIQMDLPLVPGDKIHCLDILFAFTKNVLGESGELDSLKANMEEKFMATNLSKSSYEPIATTLRWKQEDISATVIQKAYRSYVLHRSMALSNTPCVPRAEEEAASLPDEGFVAFTANENCVLPDKSETASATSFPPSYESVTRGLSDRVNMRTSSSIQNEDEATSMELIAPGP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. Plays a key role in brain, probably by regulating the moment when neurotransmitters are released in neurons. Involved in sensory perception of mechanical pain: activation in somatosensory neurons induces pain without neurogenic inflammation and produces hypersensitivity to mechanical, but not thermal stimuli.
Phosphorylation at Ser-1516 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
The S3b-S4 and S1-S2 loops of repeat IV are targeted by H.maculata toxins Hm1a and Hm1b, leading to inhibit fast inactivation of Nav1.1/SCN1A. Selectivity for H.maculata toxins Hm1a and Hm1b depends on S1-S2 loops of repeat IV.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.1/SCN1A subfamily.
Pore-forming subunit of a voltage-gated sodium channel complex through which Na(+) ions pass in accordance with their electrochemical gradient. Alternates between resting, activated and inactivated states. Required for normal muscle fiber excitability, normal muscle contraction and relaxation cycles, and constant muscle strength in the presence of fluctuating K(+) levels.
Phosphorylation at Ser-1328 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.4/SCN4A subfamily.
This protein mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-resistant Na(+) channel isoform. This channel is responsible for the initial upstroke of the action potential. Channel inactivation is regulated by intracellular calcium levels.
Ubiquitinated by NEDD4L; which promotes its endocytosis. Does not seem to be ubiquitinated by NEDD4 or WWP2.
Phosphorylation at Ser-1503 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents (Probable). Regulated through phosphorylation by CaMK2D (By similarity).
Lacks the cysteine which covalently binds the conotoxin GVIIJ. This cysteine (position 868) is speculated in other sodium channel subunits alpha to be implied in covalent binding with the sodium channel subunit beta-2 or beta-4.
Cell membrane>Multi-pass membrane protein. Cytoplasm>Perinuclear region. Cell membrane>Sarcolemma>T-tubule.
Note: RANGRF promotes trafficking to the cell membrane.
Found in jejunal circular smooth muscle cells (at protein level). Expressed in human atrial and ventricular cardiac muscle but not in adult skeletal muscle, brain, myometrium, liver, or spleen. Isoform 4 is expressed in brain.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
The IQ domain mediates association with calmodulin.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.5/SCN5A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-sensitive Na(+) channel isoform. Plays a role in pain mechanisms, especially in the development of inflammatory pain.
Phosphorylation at Ser-1490 by PKC in a highly conserved cytoplasmic loop increases peak sodium currents.
Ubiquitinated by NEDD4L; which may promote its endocytosis. Does not seem to be ubiquitinated by NEDD4.
Cell membrane>Multi-pass membrane protein. Cell projection>Neuron projection.
Note: In neurite terminals.
Expressed strongly in dorsal root ganglion, with only minor levels elsewhere in the body, smooth muscle cells, MTC cell line and C-cell carcinoma. Isoform 1 is expressed preferentially in the central and peripheral nervous system. Isoform 2 is expressed preferentially in the dorsal root ganglion.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.7/SCN9A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. Implicated in the regulation of hippocampal replay occurring within sharp wave ripples (SPW-R) important for memory (By similarity).
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1506 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Sumoylated at Lys-38. Sumoylation is induced by hypoxia, increases voltage-gated sodium current and mediates the early response to acute hypoxia in neurons. Sumoylated SCN2A is located at the cell membrane.
Cell membrane>Multi-pass membrane protein.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.2/SCN2A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient. May contribute to the regulation of serotonin/5-hydroxytryptamine release by enterochromaffin cells (By similarity). In pancreatic endocrine cells, required for both glucagon and glucose-induced insulin secretion (By similarity).
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1501 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
Expressed in enterochromaffin cells in both colon and small bowel (at protein level).
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.3/SCN3A subfamily.
This protein mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which sodium ions may pass in accordance with their electrochemical gradient. It is a tetrodotoxin-resistant sodium channel isoform. Also involved, with the contribution of the receptor tyrosine kinase NTRK2, in rapid BDNF-evoked neuronal depolarization.
Phosphorylation at Ser-1341 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein.
Expressed in the dorsal root ganglia and trigeminal ganglia, olfactory bulb, hippocampus, cerebellar cortex, spinal cord, spleen, small intestine and placenta.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.9/SCN11A subfamily.
Mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which Na(+) ions may pass in accordance with their electrochemical gradient.
In macrophages and melanoma cells, may participate in the control of podosome and invadopodia formation.
May be ubiquitinated by NEDD4L; which would promote its endocytosis.
Phosphorylation at Ser-1497 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Cell membrane>Multi-pass membrane protein. Cell projection>Axon.
Note: Mainly localizes to the axon initial segment.
Cytoplasmic vesicle.
Note: Some vesicles are localized adjacent to melanoma invadopodia and macrophage podosomes. Does not localize to the plasma membrane.
Expressed in the hippocampus with increased expression in epileptic tissue compared to normal adjacent tissue (at protein level). Isoform 5: Expressed in non-neuronal tissues, such as monocytes/macrophages.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.6/SCN8A subfamily.
Tetrodotoxin-resistant channel that mediates the voltage-dependent sodium ion permeability of excitable membranes. Assuming opened or closed conformations in response to the voltage difference across the membrane, the protein forms a sodium-selective channel through which sodium ions may pass in accordance with their electrochemical gradient. Plays a role in neuropathic pain mechanisms.
Ubiquitinated by NEDD4L; which promotes its endocytosis.
Phosphorylation at Ser-1451 by PKC in a highly conserved cytoplasmic loop slows inactivation of the sodium channel and reduces peak sodium currents.
Lacks the cysteine which covalently binds the conotoxin GVIIJ. This cysteine (position 816) is speculated in other sodium channel subunits alpha to be implied in covalent binding with the sodium channel subunit beta-2 or beta-4.
Cell membrane>Multi-pass membrane protein.
Note: It can be translocated to the cell membrane through association with S100A10.
Expressed in the dorsal root ganglia and sciatic nerve.
The sequence contains 4 internal repeats, each with 5 hydrophobic segments (S1, S2, S3, S5, S6) and one positively charged segment (S4). Segments S4 are probably the voltage-sensors and are characterized by a series of positively charged amino acids at every third position.
Belongs to the sodium channel (TC 1.A.1.10) family. Nav1.8/SCN10A subfamily.
Research Fields
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Nervous system > Dopaminergic synapse.
· Organismal Systems > Sensory system > Taste transduction.
References
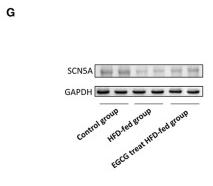
Application: WB Species: Human Sample: HepG2 cells
Application: WB Species: Rat Sample: heart
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.