NMNAT2 Antibody - #DF13581
| Product: | NMNAT2 Antibody |
| Catalog: | DF13581 |
| Description: | Rabbit polyclonal antibody to NMNAT2 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 34kDa; 34kD(Calculated). |
| Uniprot: | Q9BZQ4 |
| RRID: | AB_2846600 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF13581, RRID:AB_2846600.
Fold/Unfold
C1orf15; KIAA0479; MGC2756; NaMN adenylyltransferase 2; Nicotinamide mononucleotide adenylyltransferase 2; Nicotinamide nucleotide adenylyltransferase 2; Nicotinate-nucleotide adenylyltransferase 2; NMN adenylyltransferase 2; NMNA2_HUMAN; NMNAT 2; Nmnat2; PNAT 2; PNAT2; Pyridine nucleotide adenylyltransferase 2;
Immunogens
A synthesized peptide derived from human NMNAT2, corresponding to a region within C-terminal amino acids.
Highly expressed in brain, in particular in cerebrum, cerebellum, occipital lobe, frontal lobe, temporal lobe and putamen. Also found in the heart, skeletal muscle, pancreas and islets of Langerhans.
- Q9BZQ4 NMNA2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTETTKTHVILLACGSFNPITKGHIQMFERARDYLHKTGRFIVIGGIVSPVHDSYGKQGLVSSRHRLIMCQLAVQNSDWIRVDPWECYQDTWQTTCSVLEHHRDLMKRVTGCILSNVNTPSMTPVIGQPQNETPQPIYQNSNVATKPTAAKILGKVGESLSRICCVRPPVERFTFVDENANLGTVMRYEEIELRILLLCGSDLLESFCIPGLWNEADMEVIVGDFGIVVVPRDAADTDRIMNHSSILRKYKNNIMVVKDDINHPMSVVSSTKSRLALQHGDGHVVDYLSQPVIDYILKSQLYINASG
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Nicotinamide/nicotinate-nucleotide adenylyltransferase that acts as an axon maintenance factor (By similarity). Catalyzes the formation of NAD(+) from nicotinamide mononucleotide (NMN) and ATP. Can also use the deamidated form; nicotinic acid mononucleotide (NaMN) as substrate but with a lower efficiency. Cannot use triazofurin monophosphate (TrMP) as substrate. Also catalyzes the reverse reaction, i.e. the pyrophosphorolytic cleavage of NAD(+). For the pyrophosphorolytic activity prefers NAD(+), NADH and NaAD as substrates and degrades nicotinic acid adenine dinucleotide phosphate (NHD) less effectively. Fails to cleave phosphorylated dinucleotides NADP(+), NADPH and NaADP(+). Axon survival factor required for the maintenance of healthy axons: acts by delaying Wallerian axon degeneration, an evolutionarily conserved process that drives the loss of damaged axons (By similarity).
Degraded in response to injured neurite. Degradation is probably caused by ubiquitination by MYCBP2 (By similarity). Ubiquitinated on threonine and/or serine residues by MYCBP2; consequences of threonine and/or serine ubiquitination are however unclear.
Palmitoylated; palmitoylation is required for membrane association.
Golgi apparatus membrane>Lipid-anchor. Cytoplasmic vesicle membrane>Lipid-anchor. Cytoplasm. Cell projection>Axon.
Note: Delivered to axons with Golgi-derived cytoplasmic vesicles.
Highly expressed in brain, in particular in cerebrum, cerebellum, occipital lobe, frontal lobe, temporal lobe and putamen. Also found in the heart, skeletal muscle, pancreas and islets of Langerhans.
Belongs to the eukaryotic NMN adenylyltransferase family.
Research Fields
· Metabolism > Metabolism of cofactors and vitamins > Nicotinate and nicotinamide metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
References
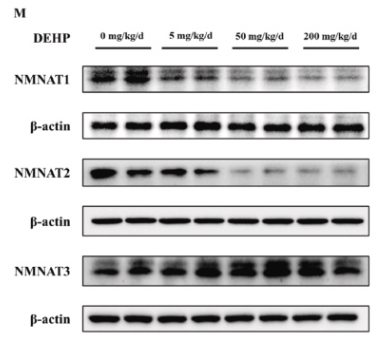
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.