Phospho-AMPK alpha 1 (Ser486) Antibody - #AF3422
| Product: | Phospho-AMPK alpha 1 (Ser486) Antibody |
| Catalog: | AF3422 |
| Description: | Rabbit polyclonal antibody to Phospho-AMPK alpha 1 (Ser486) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Zebrafish, Bovine, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 62kDa, 70 kDa; 64kD(Calculated). |
| Uniprot: | Q13131 |
| RRID: | AB_2834864 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3422, RRID:AB_2834864.
Fold/Unfold
5 AMP activated protein kinase alpha 1catalytic subunit; 5 AMP activated protein kinase catalytic alpha 1 chain; 5' AMP activated protein kinase catalytic subunit alpha 1; 5'-AMP-activated protein kinase catalytic subunit alpha-1; AAPK1; AAPK1_HUMAN; ACACA kinase; acetyl CoA carboxylase kinase; AI194361; AI450832; AL024255; AMP -activate kinase alpha 1 subunit; AMP-activated protein kinase, catalytic, alpha -1; AMPK 1; AMPK alpha 1; AMPK alpha 1 chain; AMPK; AMPK subunit alpha-1; AMPK1; AMPKa1; AMPKalpha1; C130083N04Rik; cb116; EC 2.7.11.1; HMG CoA reductase kinase; HMGCR kinase; hormone sensitive lipase kinase; Hydroxymethylglutaryl CoA reductase kinase; im:7154392; kinase AMPK alpha1; MGC33776; MGC57364; OTTHUMP00000161795; OTTHUMP00000161796; PRKAA 1; PRKAA1; Protein kinase AMP activated alpha 1 catalytic subunit; SNF1-like protein AMPK; SNF1A; Tau protein kinase PRKAA1; wu:fa94c10;
Immunogens
A synthesized peptide derived from human AMPK alpha 1 around the phosphorylation site of Ser486.
- Q13131 AAPK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MRRLSSWRKMATAEKQKHDGRVKIGHYILGDTLGVGTFGKVKVGKHELTGHKVAVKILNRQKIRSLDVVGKIRREIQNLKLFRHPHIIKLYQVISTPSDIFMVMEYVSGGELFDYICKNGRLDEKESRRLFQQILSGVDYCHRHMVVHRDLKPENVLLDAHMNAKIADFGLSNMMSDGEFLRTSCGSPNYAAPEVISGRLYAGPEVDIWSSGVILYALLCGTLPFDDDHVPTLFKKICDGIFYTPQYLNPSVISLLKHMLQVDPMKRATIKDIREHEWFKQDLPKYLFPEDPSYSSTMIDDEALKEVCEKFECSEEEVLSCLYNRNHQDPLAVAYHLIIDNRRIMNEAKDFYLATSPPDSFLDDHHLTRPHPERVPFLVAETPRARHTLDELNPQKSKHQGVRKAKWHLGIRSQSRPNDIMAEVCRAIKQLDYEWKVVNPYYLRVRRKNPVTSTYSKMSLQLYQVDSRTYLLDFRSIDDEITEAKSGTATPQRSGSVSNYRSCQRSDSDAEAQGKSSEVSLTSSVTSLDSSPVDLTPRPGSHTIEFFEMCANLIKILAQ
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Catalytic subunit of AMP-activated protein kinase (AMPK), an energy sensor protein kinase that plays a key role in regulating cellular energy metabolism. In response to reduction of intracellular ATP levels, AMPK activates energy-producing pathways and inhibits energy-consuming processes: inhibits protein, carbohydrate and lipid biosynthesis, as well as cell growth and proliferation. AMPK acts via direct phosphorylation of metabolic enzymes, and by longer-term effects via phosphorylation of transcription regulators. Also acts as a regulator of cellular polarity by remodeling the actin cytoskeleton; probably by indirectly activating myosin. Regulates lipid synthesis by phosphorylating and inactivating lipid metabolic enzymes such as ACACA, ACACB, GYS1, HMGCR and LIPE; regulates fatty acid and cholesterol synthesis by phosphorylating acetyl-CoA carboxylase (ACACA and ACACB) and hormone-sensitive lipase (LIPE) enzymes, respectively. Regulates insulin-signaling and glycolysis by phosphorylating IRS1, PFKFB2 and PFKFB3. AMPK stimulates glucose uptake in muscle by increasing the translocation of the glucose transporter SLC2A4/GLUT4 to the plasma membrane, possibly by mediating phosphorylation of TBC1D4/AS160. Regulates transcription and chromatin structure by phosphorylating transcription regulators involved in energy metabolism such as CRTC2/TORC2, FOXO3, histone H2B, HDAC5, MEF2C, MLXIPL/ChREBP, EP300, HNF4A, p53/TP53, SREBF1, SREBF2 and PPARGC1A. Acts as a key regulator of glucose homeostasis in liver by phosphorylating CRTC2/TORC2, leading to CRTC2/TORC2 sequestration in the cytoplasm. In response to stress, phosphorylates 'Ser-36' of histone H2B (H2BS36ph), leading to promote transcription. Acts as a key regulator of cell growth and proliferation by phosphorylating TSC2, RPTOR and ATG1/ULK1: in response to nutrient limitation, negatively regulates the mTORC1 complex by phosphorylating RPTOR component of the mTORC1 complex and by phosphorylating and activating TSC2. In response to nutrient limitation, promotes autophagy by phosphorylating and activating ATG1/ULK1. In that process also activates WDR45. In response to nutrient limitation, phosphorylates transcription factor FOXO3 promoting FOXO3 mitochondrial import (By similarity). AMPK also acts as a regulator of circadian rhythm by mediating phosphorylation of CRY1, leading to destabilize it. May regulate the Wnt signaling pathway by phosphorylating CTNNB1, leading to stabilize it. Also has tau-protein kinase activity: in response to amyloid beta A4 protein (APP) exposure, activated by CAMKK2, leading to phosphorylation of MAPT/TAU; however the relevance of such data remains unclear in vivo. Also phosphorylates CFTR, EEF2K, KLC1, NOS3 and SLC12A1.
Ubiquitinated.
Phosphorylated at Thr-183 by STK11/LKB1 in complex with STE20-related adapter-alpha (STRADA) pseudo kinase and CAB39. Also phosphorylated at Thr-183 by CAMKK2; triggered by a rise in intracellular calcium ions, without detectable changes in the AMP/ATP ratio. CAMKK1 can also phosphorylate Thr-183, but at a much lower level. Dephosphorylated by protein phosphatase 2A and 2C (PP2A and PP2C). Phosphorylated by ULK1 and ULK2; leading to negatively regulate AMPK activity and suggesting the existence of a regulatory feedback loop between ULK1, ULK2 and AMPK. Dephosphorylated by PPM1A and PPM1B.
Cytoplasm. Nucleus.
Note: In response to stress, recruited by p53/TP53 to specific promoters.
The AIS (autoinhibitory sequence) region shows some sequence similarity with the ubiquitin-associated domains and represses kinase activity.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. SNF1 subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Cardiovascular diseases > Hypertrophic cardiomyopathy (HCM).
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway - multiple species. (View pathway)
· Organismal Systems > Environmental adaptation > Circadian rhythm. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Adipocytokine signaling pathway.
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
· Organismal Systems > Endocrine system > Glucagon signaling pathway.
References
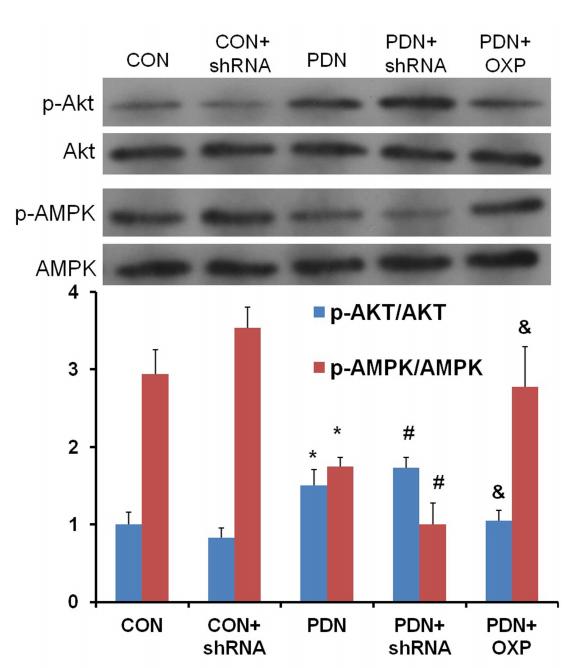
Application: WB Species: rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.