SNAP29 Antibody - #DF12743
| Product: | SNAP29 Antibody |
| Catalog: | DF12743 |
| Description: | Rabbit polyclonal antibody to SNAP29 |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Bovine, Rabbit |
| Mol.Wt.: | 29 kDa; 29kD(Calculated). |
| Uniprot: | O95721 |
| RRID: | AB_2845704 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12743, RRID:AB_2845704.
Fold/Unfold
CEDNIK; FLJ21051; SNAP 29; SNAP-29; SNAP29; SNP29_HUMAN; Soluble 29 kDa NSF attachment protein; Synaptosomal associated protein 29; Synaptosomal associated protein 29kDa; Synaptosomal-associated protein 29; Vesicle membrane fusion protein SNAP 29; Vesicle membrane fusion protein SNAP29; Vesicle-membrane fusion protein SNAP-29;
Immunogens
A synthesized peptide derived from human SNAP29, corresponding to a region within N-terminal amino acids.
Found in brain, heart, kidney, liver, lung, placenta, skeletal muscle, spleen and pancreas.
- O95721 SNP29_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSAYPKSYNPFDDDGEDEGARPAPWRDARDLPDGPDAPADRQQYLRQEVLRRAEATAASTSRSLALMYESEKVGVASSEELARQRGVLERTEKMVDKMDQDLKISQKHINSIKSVFGGLVNYFKSKPVETPPEQNGTLTSQPNNRLKEAISTSKEQEAKYQASHPNLRKLDDTDPVPRGAGSAMSTDAYPKNPHLRAYHQKIDSNLDELSMGLGRLKDIALGMQTEIEEQDDILDRLTTKVDKLDVNIKSTERKVRQL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
SNAREs, soluble N-ethylmaleimide-sensitive factor-attachment protein receptors, are essential proteins for fusion of cellular membranes. SNAREs localized on opposing membranes assemble to form a trans-SNARE complex, an extended, parallel four alpha-helical bundle that drives membrane fusion. SNAP29 is a SNARE involved in autophagy through the direct control of autophagosome membrane fusion with the lysososome membrane. Plays also a role in ciliogenesis by regulating membrane fusions.
Cytoplasm. Golgi apparatus membrane>Peripheral membrane protein. Cytoplasmic vesicle>Autophagosome membrane>Peripheral membrane protein. Cell projection>Cilium membrane>Peripheral membrane protein.
Note: Appears to be mostly membrane-bound, probably via interaction with syntaxins, but a significant portion is cytoplasmic. Localizes to the ciliary pocket from where the cilium protrudes.
Found in brain, heart, kidney, liver, lung, placenta, skeletal muscle, spleen and pancreas.
Belongs to the SNAP-25 family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Genetic Information Processing > Folding, sorting and degradation > SNARE interactions in vesicular transport.
References
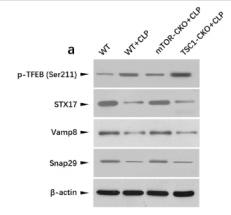
Application: WB Species: Mice Sample: CD4 + T cells
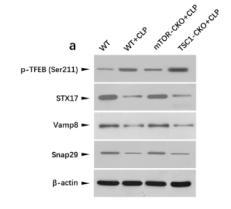
Application: WB Species: mouse Sample: CD4+T cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.