RYR2 Antibody - #AF0015
| Product: | RYR2 Antibody |
| Catalog: | AF0015 |
| Description: | Rabbit polyclonal antibody to RYR2 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit |
| Mol.Wt.: | 564 kDa; 565kD(Calculated). |
| Uniprot: | Q92736 |
| RRID: | AB_2845458 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0015, RRID:AB_2845458.
Fold/Unfold
ARVC2; ARVD2; Cardiac muscle ryanodine receptor; Cardiac muscle ryanodine receptor-calcium release channel; hRYR-2; ryanodine receptor 2 (cardiac); Ryanodine receptor 2; RyR; RYR-2; RyR2; RYR2_HUMAN; Type 2 ryanodine receptor; VTSIP;
Immunogens
A synthesized peptide derived from human RYR2, corresponding to a region within the internal amino acids.
Detected in heart muscle (at protein level). Heart muscle, brain (cerebellum and hippocampus) and placenta.
- Q92736 RYR2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MADGGEGEDEIQFLRTDDEVVLQCTATIHKEQQKLCLAAEGFGNRLCFLESTSNSKNVPPDLSICTFVLEQSLSVRALQEMLANTVEKSEGQVDVEKWKFMMKTAQGGGHRTLLYGHAILLRHSYSGMYLCCLSTSRSSTDKLAFDVGLQEDTTGEACWWTIHPASKQRSEGEKVRVGDDLILVSVSSERYLHLSYGNGSLHVDAAFQQTLWSVAPISSGSEAAQGYLIGGDVLRLLHGHMDECLTVPSGEHGEEQRRTVHYEGGAVSVHARSLWRLETLRVAWSGSHIRWGQPFRLRHVTTGKYLSLMEDKNLLLMDKEKADVKSTAFTFRSSKEKLDVGVRKEVDGMGTSEIKYGDSVCYIQHVDTGLWLTYQSVDVKSVRMGSIQRKAIMHHEGHMDDGISLSRSQHEESRTARVIRSTVFLFNRFIRGLDALSKKAKASTVDLPIESVSLSLQDLIGYFHPPDEHLEHEDKQNRLRALKNRQNLFQEEGMINLVLECIDRLHVYSSAAHFADVAGREAGESWKSILNSLYELLAALIRGNRKNCAQFSGSLDWLISRLERLEASSGILEVLHCVLVESPEALNIIKEGHIKSIISLLDKHGRNHKVLDVLCSLCVCHGVAVRSNQHLICDNLLPGRDLLLQTRLVNHVSSMRPNIFLGVSEGSAQYKKWYYELMVDHTEPFVTAEATHLRVGWASTEGYSPYPGGGEEWGGNGVGDDLFSYGFDGLHLWSGCIARTVSSPNQHLLRTDDVISCCLDLSAPSISFRINGQPVQGMFENFNIDGLFFPVVSFSAGIKVRFLLGGRHGEFKFLPPPGYAPCYEAVLPKEKLKVEHSREYKQERTYTRDLLGPTVSLTQAAFTPIPVDTSQIVLPPHLERIREKLAENIHELWVMNKIELGWQYGPVRDDNKRQHPCLVEFSKLPEQERNYNLQMSLETLKTLLALGCHVGISDEHAEDKVKKMKLPKNYQLTSGYKPAPMDLSFIKLTPSQEAMVDKLAENAHNVWARDRIRQGWTYGIQQDVKNRRNPRLVPYTLLDDRTKKSNKDSLREAVRTLLGYGYNLEAPDQDHAARAEVCSGTGERFRIFRAEKTYAVKAGRWYFEFETVTAGDMRVGWSRPGCQPDQELGSDERAFAFDGFKAQRWHQGNEHYGRSWQAGDVVGCMVDMNEHTMMFTLNGEILLDDSGSELAFKDFDVGDGFIPVCSLGVAQVGRMNFGKDVSTLKYFTICGLQEGYEPFAVNTNRDITMWLSKRLPQFLQVPSNHEHIEVTRIDGTIDSSPCLKVTQKSFGSQNSNTDIMFYRLSMPIECAEVFSKTVAGGLPGAGLFGPKNDLEDYDADSDFEVLMKTAHGHLVPDRVDKDKEATKPEFNNHKDYAQEKPSRLKQRFLLRRTKPDYSTSHSARLTEDVLADDRDDYDFLMQTSTYYYSVRIFPGQEPANVWVGWITSDFHQYDTGFDLDRVRTVTVTLGDEKGKVHESIKRSNCYMVCAGESMSPGQGRNNNGLEIGCVVDAASGLLTFIANGKELSTYYQVEPSTKLFPAVFAQATSPNVFQFELGRIKNVMPLSAGLFKSEHKNPVPQCPPRLHVQFLSHVLWSRMPNQFLKVDVSRISERQGWLVQCLDPLQFMSLHIPEENRSVDILELTEQEELLKFHYHTLRLYSAVCALGNHRVAHALCSHVDEPQLLYAIENKYMPGLLRAGYYDLLIDIHLSSYATARLMMNNEYIVPMTEETKSITLFPDENKKHGLPGIGLSTSLRPRMQFSSPSFVSISNECYQYSPEFPLDILKSKTIQMLTEAVKEGSLHARDPVGGTTEFLFVPLIKLFYTLLIMGIFHNEDLKHILQLIEPSVFKEAATPEEESDTLEKELSVDDAKLQGAGEEEAKGGKRPKEGLLQMKLPEPVKLQMCLLLQYLCDCQVRHRIEAIVAFSDDFVAKLQDNQRFRYNEVMQALNMSAALTARKTKEFRSPPQEQINMLLNFKDDKSECPCPEEIRDQLLDFHEDLMTHCGIELDEDGSLDGNSDLTIRGRLLSLVEKVTYLKKKQAEKPVESDSKKSSTLQQLISETMVRWAQESVIEDPELVRAMFVLLHRQYDGIGGLVRALPKTYTINGVSVEDTINLLASLGQIRSLLSVRMGKEEEKLMIRGLGDIMNNKVFYQHPNLMRALGMHETVMEVMVNVLGGGESKEITFPKMVANCCRFLCYFCRISRQNQKAMFDHLSYLLENSSVGLASPAMRGSTPLDVAAASVMDNNELALALREPDLEKVVRYLAGCGLQSCQMLVSKGYPDIGWNPVEGERYLDFLRFAVFCNGESVEENANVVVRLLIRRPECFGPALRGEGGNGLLAAMEEAIKIAEDPSRDGPSPNSGSSKTLDTEEEEDDTIHMGNAIMTFYSALIDLLGRCAPEMHLIHAGKGEAIRIRSILRSLIPLGDLVGVISIAFQMPTIAKDGNVVEPDMSAGFCPDHKAAMVLFLDRVYGIEVQDFLLHLLEVGFLPDLRAAASLDTAALSATDMALALNRYLCTAVLPLLTRCAPLFAGTEHHASLIDSLLHTVYRLSKGCSLTKAQRDSIEVCLLSICGQLRPSMMQHLLRRLVFDVPLLNEHAKMPLKLLTNHYERCWKYYCLPGGWGNFGAASEEELHLSRKLFWGIFDALSQKKYEQELFKLALPCLSAVAGALPPDYMESNYVSMMEKQSSMDSEGNFNPQPVDTSNITIPEKLEYFINKYAEHSHDKWSMDKLANGWIYGEIYSDSSKVQPLMKPYKLLSEKEKEIYRWPIKESLKTMLAWGWRIERTREGDSMALYNRTRRISQTSQVSVDAAHGYSPRAIDMSNVTLSRDLHAMAEMMAENYHNIWAKKKKMELESKGGGNHPLLVPYDTLTAKEKAKDREKAQDILKFLQINGYAVSRGFKDLELDTPSIEKRFAYSFLQQLIRYVDEAHQYILEFDGGSRGKGEHFPYEQEIKFFAKVVLPLIDQYFKNHRLYFLSAASRPLCSGGHASNKEKEMVTSLFCKLGVLVRHRISLFGNDATSIVNCLHILGQTLDARTVMKTGLESVKSALRAFLDNAAEDLEKTMENLKQGQFTHTRNQPKGVTQIINYTTVALLPMLSSLFEHIGQHQFGEDLILEDVQVSCYRILTSLYALGTSKSIYVERQRSALGECLAAFAGAFPVAFLETHLDKHNIYSIYNTKSSRERAALSLPTNVEDVCPNIPSLEKLMEEIVELAESGIRYTQMPHVMEVILPMLCSYMSRWWEHGPENNPERAEMCCTALNSEHMNTLLGNILKIIYNNLGIDEGAWMKRLAVFSQPIINKVKPQLLKTHFLPLMEKLKKKAATVVSEEDHLKAEARGDMSEAELLILDEFTTLARDLYAFYPLLIRFVDYNRAKWLKEPNPEAEELFRMVAEVFIYWSKSHNFKREEQNFVVQNEINNMSFLITDTKSKMSKAAVSDQERKKMKRKGDRYSMQTSLIVAALKRLLPIGLNICAPGDQELIALAKNRFSLKDTEDEVRDIIRSNIHLQGKLEDPAIRWQMALYKDLPNRTDDTSDPEKTVERVLDIANVLFHLEQKSKRVGRRHYCLVEHPQRSKKAVWHKLLSKQRKRAVVACFRMAPLYNLPRHRAVNLFLQGYEKSWIETEEHYFEDKLIEDLAKPGAEPPEEDEGTKRVDPLHQLILLFSRTALTEKCKLEEDFLYMAYADIMAKSCHDEEDDDGEEEVKSFEEKEMEKQKLLYQQARLHDRGAAEMVLQTISASKGETGPMVAATLKLGIAILNGGNSTVQQKMLDYLKEKKDVGFFQSLAGLMQSCSVLDLNAFERQNKAEGLGMVTEEGSGEKVLQDDEFTCDLFRFLQLLCEGHNSDFQNYLRTQTGNNTTVNIIISTVDYLLRVQESISDFYWYYSGKDVIDEQGQRNFSKAIQVAKQVFNTLTEYIQGPCTGNQQSLAHSRLWDAVVGFLHVFAHMQMKLSQDSSQIELLKELMDLQKDMVVMLLSMLEGNVVNGTIGKQMVDMLVESSNNVEMILKFFDMFLKLKDLTSSDTFKEYDPDGKGVISKRDFHKAMESHKHYTQSETEFLLSCAETDENETLDYEEFVKRFHEPAKDIGFNVAVLLTNLSEHMPNDTRLQTFLELAESVLNYFQPFLGRIEIMGSAKRIERVYFEISESSRTQWEKPQVKESKRQFIFDVVNEGGEKEKMELFVNFCEDTIFEMQLAAQISESDLNERSANKEESEKERPEEQGPRMAFFSILTVRSALFALRYNILTLMRMLSLKSLKKQMKKVKKMTVKDMVTAFFSSYWSIFMTLLHFVASVFRGFFRIICSLLLGGSLVEGAKKIKVAELLANMPDPTQDEVRGDGEEGERKPLEAALPSEDLTDLKELTEESDLLSDIFGLDLKREGGQYKLIPHNPNAGLSDLMSNPVPMPEVQEKFQEQKAKEEEKEEKEETKSEPEKAEGEDGEKEEKAKEDKGKQKLRQLHTHRYGEPEVPESAFWKKIIAYQQKLLNYFARNFYNMRMLALFVAFAINFILLFYKVSTSSVVEGKELPTRSSSENAKVTSLDSSSHRIIAVHYVLEESSGYMEPTLRILAILHTVISFFCIIGYYCLKVPLVIFKREKEVARKLEFDGLYITEQPSEDDIKGQWDRLVINTQSFPNNYWDKFVKRKVMDKYGEFYGRDRISELLGMDKAALDFSDAREKKKPKKDSSLSAVLNSIDVKYQMWKLGVVFTDNSFLYLAWYMTMSVLGHYNNFFFAAHLLDIAMGFKTLRTILSSVTHNGKQLVLTVGLLAVVVYLYTVVAFNFFRKFYNKSEDGDTPDMKCDDMLTCYMFHMYVGVRAGGGIGDEIEDPAGDEYEIYRIIFDITFFFFVIVILLAIIQGLIIDAFGELRDQQEQVKEDMETKCFICGIGNDYFDTVPHGFETHTLQEHNLANYLFFLMYLINKDETEHTGQESYVWKMYQERCWEFFPAGDCFRKQYEDQLN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Calcium channel that mediates the release of Ca(2+) from the sarcoplasmic reticulum into the cytoplasm and thereby plays a key role in triggering cardiac muscle contraction. Aberrant channel activation can lead to cardiac arrhythmia. In cardiac myocytes, calcium release is triggered by increased Ca(2+) levels due to activation of the L-type calcium channel CACNA1C. The calcium channel activity is modulated by formation of heterotetramers with RYR3. Required for cellular calcium ion homeostasis. Required for embryonic heart development.
Channel activity is modulated by phosphorylation. Phosphorylation at Ser-2808 and Ser-2814 increases the open probability of the calcium channel. Phosphorylation is increased in failing heart, leading to calcium leaks and increased cytoplasmic Ca(2+) levels.
Phosphorylation at Ser-2031 by PKA enhances the response to lumenal calcium.
Sarcoplasmic reticulum membrane>Multi-pass membrane protein. Membrane>Multi-pass membrane protein. Sarcoplasmic reticulum.
Note: The number of predicted transmembrane domains varies between orthologs, but both N-terminus and C-terminus seem to be cytoplasmic.
Detected in heart muscle (at protein level). Heart muscle, brain (cerebellum and hippocampus) and placenta.
The calcium release channel activity resides in the C-terminal region while the remaining part of the protein resides in the cytoplasm.
Belongs to the ryanodine receptor (TC 1.A.3.1) family. RYR2 subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > Calcium signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Cardiovascular diseases > Hypertrophic cardiomyopathy (HCM).
· Human Diseases > Cardiovascular diseases > Arrhythmogenic right ventricular cardiomyopathy (ARVC).
· Human Diseases > Cardiovascular diseases > Dilated cardiomyopathy (DCM).
· Organismal Systems > Circulatory system > Cardiac muscle contraction. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Environmental adaptation > Circadian entrainment.
· Organismal Systems > Endocrine system > Insulin secretion. (View pathway)
· Organismal Systems > Endocrine system > Oxytocin signaling pathway.
· Organismal Systems > Digestive system > Pancreatic secretion.
References
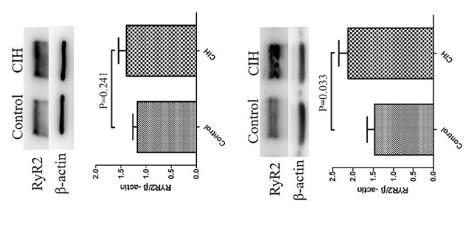
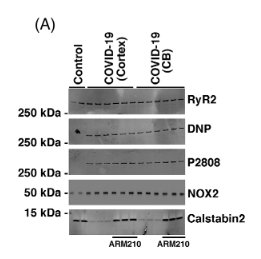
Application: WB Species: Human Sample: brain
Application: WB Species: rat Sample: atrial
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.