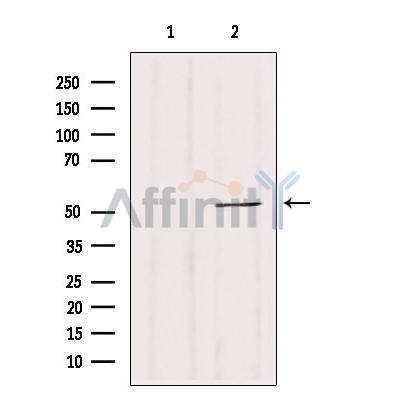
CALCOCO2 Antibody - #DF12360
| Product: | CALCOCO2 Antibody |
| Catalog: | DF12360 |
| Description: | Rabbit polyclonal antibody to CALCOCO2 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 52 kDa; 52kD(Calculated). |
| Uniprot: | Q13137 |
| RRID: | AB_2845165 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12360, RRID:AB_2845165.
Fold/Unfold
Antigen nuclear dot 52 kDa protein; CACO2_HUMAN; Calcium binding and coiled coil domain 2; Calcium binding and coiled coil domain containing protein 2; Calcium-binding and coiled-coil domain-containing protein 2; CALCOCO 2; CALCOCO2; MGC17318; NDP52; Nuclear domain 10 protein 52; Nuclear domain 10 protein; Nuclear domain 10 protein NDP52; Nuclear domain protein 52; Nuclear dot protein 52; OTTHUMP00000218344; OTTHUMP00000218345; OTTHUMP00000218346; OTTHUMP00000218347; OTTHUMP00000218349; OTTHUMP00000218351;
Immunogens
Expressed in all tissues tested with highest expression in skeletal muscle and lowest in brain.
- Q13137 CACO2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEETIKDPPTSAVLLDHCHFSQVIFNSVEKFYIPGGDVTCHYTFTQHFIPRRKDWIGIFRVGWKTTREYYTFMWVTLPIDLNNKSAKQQEVQFKAYYLPKDDEYYQFCYVDEDGVVRGASIPFQFRPENEEDILVVTTQGEVEEIEQHNKELCKENQELKDSCISLQKQNSDMQAELQKKQEELETLQSINKKLELKVKEQKDYWETELLQLKEQNQKMSSENEKMGIRVDQLQAQLSTQEKEMEKLVQGDQDKTEQLEQLKKENDHLFLSLTEQRKDQKKLEQTVEQMKQNETTAMKKQQELMDENFDLSKRLSENEIICNALQRQKERLEGENDLLKRENSRLLSYMGLDFNSLPYQVPTSDEGGARQNPGLAYGNPYSGIQESSSPSPLSIKKCPICKADDICDHTLEQQQMQPLCFNCPICDKIFPATEKQIFEDHVFCHSL
PTMs - Q13137 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K87 | Ubiquitination | Uniprot | |
| K94 | Ubiquitination | Uniprot | |
| Y96 | Phosphorylation | Uniprot | |
| K160 | Ubiquitination | Uniprot | |
| K168 | Ubiquitination | Uniprot | |
| K180 | Ubiquitination | Uniprot | |
| K192 | Ubiquitination | Uniprot | |
| Y204 | Phosphorylation | Uniprot | |
| K213 | Ubiquitination | Uniprot | |
| K225 | Ubiquitination | Uniprot | |
| S238 | Phosphorylation | Uniprot | |
| K242 | Ubiquitination | Uniprot | |
| K262 | Ubiquitination | Uniprot | |
| S271 | Phosphorylation | Uniprot | |
| K281 | Ubiquitination | Uniprot | |
| K299 | Ubiquitination | Uniprot | |
| K312 | Ubiquitination | Uniprot | |
| S315 | Phosphorylation | Uniprot | |
| K339 | Ubiquitination | Uniprot | |
| S347 | Phosphorylation | Uniprot | |
| S355 | Phosphorylation | Uniprot | |
| Y358 | Phosphorylation | Uniprot | |
| Y376 | Phosphorylation | Uniprot | |
| Y380 | Phosphorylation | Uniprot | |
| S381 | Phosphorylation | Uniprot | |
| S387 | Phosphorylation | Uniprot | |
| S388 | Phosphorylation | Uniprot | |
| S390 | Phosphorylation | Uniprot | |
| K395 | Ubiquitination | Uniprot | |
| K396 | Ubiquitination | Uniprot | |
| K401 | Ubiquitination | Uniprot | |
| S445 | Phosphorylation | Uniprot |
Research Backgrounds
Xenophagy-specific receptor required for autophagy-mediated intracellular bacteria degradation. Acts as an effector protein of galectin-sensed membrane damage that restricts the proliferation of infecting pathogens such as Salmonella typhimurium upon entry into the cytosol by targeting LGALS8-associated bacteria for autophagy. Initially orchestrates bacteria targeting to autophagosomes and subsequently ensures pathogen degradation by regulating pathogen-containing autophagosome maturation. Bacteria targeting to autophagosomes relies on its interaction with MAP1LC3A, MAP1LC3B and/or GABARAPL2, whereas regulation of pathogen-containing autophagosome maturation requires the interaction with MAP3LC3C. May play a role in ruffle formation and actin cytoskeleton organization and seems to negatively regulate constitutive secretion.
Cytoplasm>Perinuclear region. Cytoplasm>Cytoskeleton. Cytoplasmic vesicle>Autophagosome membrane>Peripheral membrane protein.
Note: According to PubMed:7540613, localizes to nuclear dots. According to PubMed:9230084 and PubMed:12869526, it is not a nuclear dot-associated protein but localizes predominantly in the cytoplasm with a coarse-grained distribution preferentially close to the nucleus.
Expressed in all tissues tested with highest expression in skeletal muscle and lowest in brain.
Dimer. Part of a complex consisting of CALCOCO2, TAX1BP1 and MYO6. Interacts with GEMIN4. Interacts with ATG8 family members MAP1LC3A, MAP1LC3B, GABARAP, GABARAPL1 and GABARAPL2. Interacts with ATG8 family member MAP1LC3C. Interacts with LGALS8.
The MYO6-binding domain is required for autophagy-mediated degradation of infecting bacteria such as Salmonella typhimurium, but not for bacteria targeting to autophagosomes.
The CLIR (LC3C-interacting region) motif is required for interaction with MAP1LC3C, but dispensable for CALCOCO2-mediated autophagosome maturation.
The LIR-like motif is required for interaction with MAP1LC3A, MAP1LC3B and GABARAPL2, as well as for CALCOCO2-mediated autophagosome maturation.
The LGALS8-binding domain is essential for the recruitment to cytosol-exposed infecting bacteria.
Belongs to the CALCOCO family.
References
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.