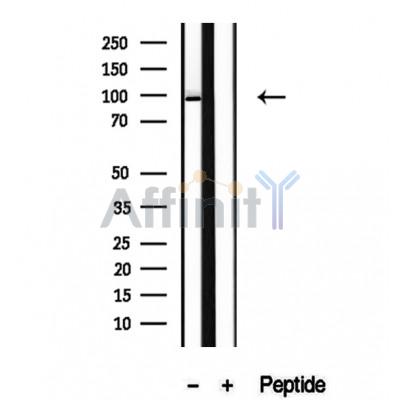
AGO2 Antibody - #DF12246
| Product: | AGO2 Antibody |
| Catalog: | DF12246 |
| Description: | Rabbit polyclonal antibody to AGO2 |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Xenopus |
| Mol.Wt.: | 90-95 kDa; 97kD(Calculated). |
| Uniprot: | Q9UKV8 |
| RRID: | AB_2845051 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12246, RRID:AB_2845051.
Fold/Unfold
Ago 2; AGO2_HUMAN; Argonaute 2; argonaute 2, RISC catalytic component; Argonaute RISC catalytic component 2; Argonaute2; CTA-204B4.6; dAgo2; eIF 2C 2; eIF-2C 2; eIF2C 2; Eif2c2; Eukaryotic translation initiation factor 2C 2; Eukaryotic translation initiation factor 2C subunit 2; hAgo2; MGC3183; PAZ Piwi domain protein; PPD; Protein argonaute-2; Protein slicer; Q10; Slicer protein;
Immunogens
- Q9UKV8 AGO2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MYSGAGPALAPPAPPPPIQGYAFKPPPRPDFGTSGRTIKLQANFFEMDIPKIDIYHYELDIKPEKCPRRVNREIVEHMVQHFKTQIFGDRKPVFDGRKNLYTAMPLPIGRDKVELEVTLPGEGKDRIFKVSIKWVSCVSLQALHDALSGRLPSVPFETIQALDVVMRHLPSMRYTPVGRSFFTASEGCSNPLGGGREVWFGFHQSVRPSLWKMMLNIDVSATAFYKAQPVIEFVCEVLDFKSIEEQQKPLTDSQRVKFTKEIKGLKVEITHCGQMKRKYRVCNVTRRPASHQTFPLQQESGQTVECTVAQYFKDRHKLVLRYPHLPCLQVGQEQKHTYLPLEVCNIVAGQRCIKKLTDNQTSTMIRATARSAPDRQEEISKLMRSASFNTDPYVREFGIMVKDEMTDVTGRVLQPPSILYGGRNKAIATPVQGVWDMRNKQFHTGIEIKVWAIACFAPQRQCTEVHLKSFTEQLRKISRDAGMPIQGQPCFCKYAQGADSVEPMFRHLKNTYAGLQLVVVILPGKTPVYAEVKRVGDTVLGMATQCVQMKNVQRTTPQTLSNLCLKINVKLGGVNNILLPQGRPPVFQQPVIFLGADVTHPPAGDGKKPSIAAVVGSMDAHPNRYCATVRVQQHRQEIIQDLAAMVRELLIQFYKSTRFKPTRIIFYRDGVSEGQFQQVLHHELLAIREACIKLEKDYQPGITFIVVQKRHHTRLFCTDKNERVGKSGNIPAGTTVDTKITHPTEFDFYLCSHAGIQGTSRPSHYHVLWDDNRFSSDELQILTYQLCHTYVRCTRSVSIPAPAYYAHLVAFRARYHLVDKEHDSAEGSHTSGQSNGRDHQALAKAVQVHQDTLRTMYFA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q9UKV8 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K62 | Ubiquitination | Uniprot | |
| K91 | Ubiquitination | Uniprot | |
| S153 | Phosphorylation | Uniprot | |
| C188 | S-Nitrosylation | Uniprot | |
| K248 | Ubiquitination | Uniprot | |
| S253 | Phosphorylation | Uniprot | |
| T303 | Phosphorylation | Uniprot | |
| T307 | Phosphorylation | Uniprot | |
| K317 | Acetylation | Uniprot | |
| K335 | Ubiquitination | Uniprot | |
| T337 | Phosphorylation | Uniprot | |
| Y338 | Phosphorylation | Uniprot | |
| T357 | Phosphorylation | Uniprot | |
| K381 | Ubiquitination | Uniprot | |
| S385 | Phosphorylation | Uniprot | |
| S387 | Phosphorylation | P49137 (MAPKAPK2) , Q9Y243 (AKT3) | Uniprot |
| T390 | Phosphorylation | Uniprot | |
| Y393 | Phosphorylation | P00533 (EGFR) | Uniprot |
| K402 | Sumoylation | Uniprot | |
| K425 | Ubiquitination | Uniprot | |
| K440 | Ubiquitination | Uniprot | |
| K468 | Ubiquitination | Uniprot | |
| K493 | Ubiquitination | Uniprot | |
| T526 | Phosphorylation | Uniprot | |
| Y529 | Phosphorylation | Uniprot | |
| K533 | Ubiquitination | Uniprot | |
| K550 | Ubiquitination | Uniprot | |
| K566 | Ubiquitination | Uniprot | |
| K655 | Ubiquitination | Uniprot | |
| K720 | Ubiquitination | Uniprot | |
| K726 | Ubiquitination | Uniprot | |
| Y749 | Phosphorylation | Uniprot | |
| S752 | Phosphorylation | Uniprot | |
| T759 | Phosphorylation | Uniprot | |
| S760 | Phosphorylation | Uniprot | |
| S798 | Phosphorylation | Uniprot | |
| S824 | Phosphorylation | Uniprot | |
| S828 | Phosphorylation | Uniprot | |
| T830 | Phosphorylation | Uniprot | |
| S831 | Phosphorylation | Uniprot | |
| S834 | Phosphorylation | Uniprot | |
| K844 | Ubiquitination | Uniprot | |
| T852 | Phosphorylation | Uniprot |
Research Backgrounds
Required for RNA-mediated gene silencing (RNAi) by the RNA-induced silencing complex (RISC). The 'minimal RISC' appears to include AGO2 bound to a short guide RNA such as a microRNA (miRNA) or short interfering RNA (siRNA). These guide RNAs direct RISC to complementary mRNAs that are targets for RISC-mediated gene silencing. The precise mechanism of gene silencing depends on the degree of complementarity between the miRNA or siRNA and its target. Binding of RISC to a perfectly complementary mRNA generally results in silencing due to endonucleolytic cleavage of the mRNA specifically by AGO2. Binding of RISC to a partially complementary mRNA results in silencing through inhibition of translation, and this is independent of endonuclease activity. May inhibit translation initiation by binding to the 7-methylguanosine cap, thereby preventing the recruitment of the translation initiation factor eIF4-E. May also inhibit translation initiation via interaction with EIF6, which itself binds to the 60S ribosomal subunit and prevents its association with the 40S ribosomal subunit. The inhibition of translational initiation leads to the accumulation of the affected mRNA in cytoplasmic processing bodies (P-bodies), where mRNA degradation may subsequently occur. In some cases RISC-mediated translational repression is also observed for miRNAs that perfectly match the 3' untranslated region (3'-UTR). Can also up-regulate the translation of specific mRNAs under certain growth conditions. Binds to the AU element of the 3'-UTR of the TNF (TNF-alpha) mRNA and up-regulates translation under conditions of serum starvation. Also required for transcriptional gene silencing (TGS), in which short RNAs known as antigene RNAs or agRNAs direct the transcriptional repression of complementary promoter regions.
Hydroxylated. 4-hydroxylation appears to enhance protein stability but is not required for miRNA-binding or endonuclease activity.
Cytoplasm>P-body. Nucleus.
Note: Translational repression of mRNAs results in their recruitment to P-bodies. Translocation to the nucleus requires IMP8.
Interacts with DICER1 through its Piwi domain and with TARBP2 during assembly of the RNA-induced silencing complex (RISC). Together, DICER1, AGO2 and TARBP2 constitute the trimeric RISC loading complex (RLC), or micro-RNA (miRNA) loading complex (miRLC). Within the RLC/miRLC, DICER1 and TARBP2 are required to process precursor miRNAs (pre-miRNAs) to mature miRNAs and then load them onto AGO2. AGO2 bound to the mature miRNA constitutes the minimal RISC and may subsequently dissociate from DICER1 and TARBP2. Note however that the term RISC has also been used to describe the trimeric RLC/miRLC. The formation of RISC complexes containing siRNAs rather than miRNAs appears to occur independently of DICER1. Interacts with AGO1. Also interacts with DDB1, DDX5, DDX6, DDX20, DHX30, DHX36, DDX47, DHX9, ELAVL, FXR1, GEMIN4, HNRNPF, IGF2BP1, ILF3, IMP8, MATR3, PABPC1, PRMT5, P4HA1, P4HB, RBM4, SART3, TNRC6A, TNRC6B, UPF1 and YBX1. Interacts with the P-body components DCP1A and XRN1. Associates with polysomes and messenger ribonucleoproteins (mNRPs). Interacts with RBM4; the interaction is modulated under stress-induced conditions, occurs under both cell proliferation and differentiation conditions and in an RNA- and phosphorylation-independent manner. Interacts with LIMD1, WTIP and AJUBA. Interacts with TRIM71; the interaction increases in presence of RNA. Interacts with APOBEC3G in an RNA-dependent manner. Interacts with APOBEC3A, APOBEC3C, APOBEC3F and APOBEC3H. Interacts with DICER1, TARBP2, EIF6, MOV10 and RPL7A (60S ribosome subunit); they form a large RNA-induced silencing complex (RISC). Interacts with FMR1. Interacts with ZFP36. Found in a complex, composed of AGO2, CHD7 and FAM172A (By similarity). Interacts with RC3H1; the interaction is RNA independent. Interacts with SND1. Interacts with SYT11 (By similarity). Interacts with CLNK.
The Piwi domain may perform RNA cleavage by a mechanism similar to that of RNase H. However, while RNase H utilizes a triad of Asp-Asp-Glu (DDE) for metal ion coordination, this protein appears to utilize a triad of Asp-Asp-His (DDH).
Belongs to the argonaute family. Ago subfamily.
References
Application: IF/ICC Species: Human Sample: OS cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.