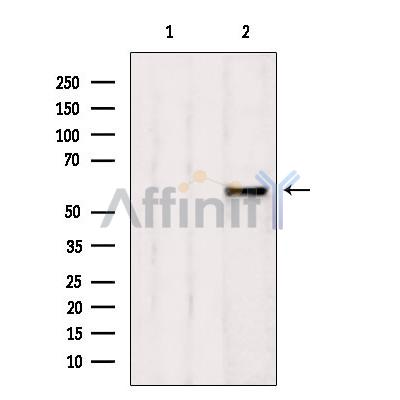
AHCYL1 Antibody - #DF12091
| Product: | AHCYL1 Antibody |
| Catalog: | DF12091 |
| Description: | Rabbit polyclonal antibody to AHCYL1 |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Mol.Wt.: | 60 kDa; 59kD(Calculated). |
| Uniprot: | O43865 |
| RRID: | AB_2844896 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF12091, RRID:AB_2844896.
Fold/Unfold
AA409031; AdoHcyase 2; Ahcy-rs3; AHCYL1; DC expressed AHCY like molecule; DC-expressed AHCY-like molecule; DCAL; Dendritic cell expressed AHCY like protein; Inositol 1,4,5-trisphosphate receptor-binding protein; IP3R binding protein released with inositol 1,4,5-trisphosphate; IRBIT; PRO0233; Putative adenosylhomocysteinase 2; S adenosyl L homocysteine hydrolase 2; S adenosylhomocysteine hydrolase like 1; S-adenosyl-L-homocysteine hydrolase 2; S-adenosylhomocysteine hydrolase-like protein 1; SAHH2_HUMAN; XPVKONA;
Immunogens
A synthesized peptide derived from human AHCYL1, corresponding to a region within N-terminal amino acids.
- O43865 SAHH2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSMPDAMPLPGVGEELKQAKEIEDAEKYSFMATVTKAPKKQIQFADDMQEFTKFPTKTGRRSLSRSISQSSTDSYSSAASYTDSSDDEVSPREKQQTNSKGSSNFCVKNIKQAEFGRREIEIAEQDMSALISLRKRAQGEKPLAGAKIVGCTHITAQTAVLIETLCALGAQCRWSACNIYSTQNEVAAALAEAGVAVFAWKGESEDDFWWCIDRCVNMDGWQANMILDDGGDLTHWVYKKYPNVFKKIRGIVEESVTGVHRLYQLSKAGKLCVPAMNVNDSVTKQKFDNLYCCRESILDGLKRTTDVMFGGKQVVVCGYGEVGKGCCAALKALGAIVYITEIDPICALQACMDGFRVVKLNEVIRQVDVVITCTGNKNVVTREHLDRMKNSCIVCNMGHSNTEIDVTSLRTPELTWERVRSQVDHVIWPDGKRVVLLAEGRLLNLSCSTVPTFVLSITATTQALALIELYNAPEGRYKQDVYLLPKKMDEYVASLHLPSFDAHLTELTDDQAKYLGLNKNGPFKPNYYRY
Research Backgrounds
Multifaceted cellular regulator which coordinates several essential cellular functions including regulation of epithelial HCO3(-) and fluid secretion, mRNA processing and DNA replication. Regulates ITPR1 sensitivity to inositol 1,4,5-trisphosphate competing for the common binding site and acting as endogenous 'pseudoligand' whose inhibitory activity can be modulated by its phosphorylation status. In the pancreatic and salivary ducts, at resting state, attenuates inositol 1,4,5-trisphosphate-induced calcium release by interacting with ITPR1. When extracellular stimuli induce ITPR1 phosphorylation or inositol 1,4,5-trisphosphate production, dissociates of ITPR1 to interact with CFTR and SLC26A6 mediating their synergistic activation by calcium and cAMP that stimulates the epithelial secretion of electrolytes and fluid (By similarity). Also activates basolateral SLC4A4 isoform 1 to coordinate fluid and HCO3(-) secretion. Inhibits the effect of STK39 on SLC4A4 and CFTR by recruiting PP1 phosphatase which activates SLC4A4, SLC26A6 and CFTR through dephosphorylation (By similarity). Mediates the induction of SLC9A3 surface expression produced by Angiotensin-2. Depending on the cell type, activates SLC9A3 in response to calcium or reverses SLC9A3R2-dependent calcium inhibition. May modulate the polyadenylation state of specific mRNAs, both by controlling the subcellular location of FIP1L1 and by inhibiting PAPOLA activity, in response to a stimulus that alters its phosphorylation state. Acts as a (dATP)-dependent inhibitor of ribonucleotide reductase large subunit RRM1, controlling the endogenous dNTP pool and ensuring normal cell cycle progression. In vitro does not exhibit any S-adenosyl-L-homocysteine hydrolase activity (By similarity).
Phosphorylated at Ser/Thr residues between Ser-68 and Thr-72 in the PEST region: required for interaction with dATP-bound RRM1 and ITPR1. Phosphorylation at Ser-68 by PRKD1 and CAMK4 is required for further phosphorylations by CSNK1A1. Phosphorylation is induced by oxidative stress. Probably phosphorylated by CAMK2A; phosphorylation at Ser-68 may be required for interaction with SLC9A3.
Endoplasmic reticulum. Cytoplasm>Cytosol. Microsome. Apical cell membrane.
Note: Associates with membranes when phosphorylated, probably through interaction with ITPR1.
Expressed in dendritic cells.
The PEST region is essential for the interaction with ITPR1, and, when phosphorylated, is also the RRM1-binding region. The PDZ-binding region is required for maximal interaction with ITPR1 and is also responsible for the IP3-insensitive interaction with ITPR1 (By similarity).
Belongs to the adenosylhomocysteinase family.
Research Fields
· Metabolism > Amino acid metabolism > Cysteine and methionine metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.