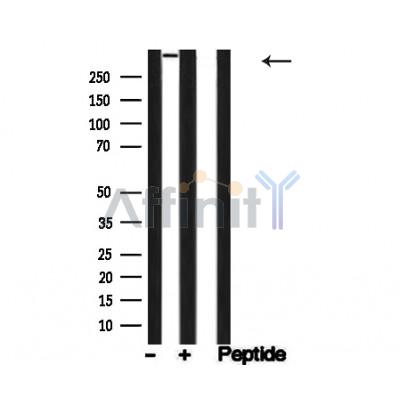
53BP1 Antibody - #AF4760
| Product: | 53BP1 Antibody |
| Catalog: | AF4760 |
| Description: | Rabbit polyclonal antibody to 53BP1 |
| Application: | WB IF/ICC |
| Reactivity: | Human, Rat |
| Prediction: | Pig, Zebrafish, Horse, Sheep, Dog, Chicken, Xenopus |
| Mol.Wt.: | 450kDa; 214kD(Calculated). |
| Uniprot: | Q12888 |
| RRID: | AB_2844754 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF4760, RRID:AB_2844754.
Fold/Unfold
53 BP1; 53BP1; FLJ41424; MGC138366; p202; p53 binding protein 1; p53 BP1; p53-binding protein 1; p53BP1; TP53 BP1; TP53B_HUMAN; Tp53bp1; TRP53 BP1; Tumor protein 53 binding protein 1; Tumor protein p53 binding protein 1; Tumor suppressor p53 binding protein 1; Tumor suppressor p53-binding protein 1;
Immunogens
A synthesized peptide derived from human 53BP1, corresponding to a region within the internal amino acids.
- Q12888 TP53B_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDPTGSQLDSDFSQQDTPCLIIEDSQPESQVLEDDSGSHFSMLSRHLPNLQTHKENPVLDVVSNPEQTAGEERGDGNSGFNEHLKENKVADPVDSSNLDTCGSISQVIEQLPQPNRTSSVLGMSVESAPAVEEEKGEELEQKEKEKEEDTSGNTTHSLGAEDTASSQLGFGVLELSQSQDVEENTVPYEVDKEQLQSVTTNSGYTRLSDVDANTAIKHEEQSNEDIPIAEQSSKDIPVTAQPSKDVHVVKEQNPPPARSEDMPFSPKASVAAMEAKEQLSAQELMESGLQIQKSPEPEVLSTQEDLFDQSNKTVSSDGCSTPSREEGGCSLASTPATTLHLLQLSGQRSLVQDSLSTNSSDLVAPSPDAFRSTPFIVPSSPTEQEGRQDKPMDTSVLSEEGGEPFQKKLQSGEPVELENPPLLPESTVSPQASTPISQSTPVFPPGSLPIPSQPQFSHDIFIPSPSLEEQSNDGKKDGDMHSSSLTVECSKTSEIEPKNSPEDLGLSLTGDSCKLMLSTSEYSQSPKMESLSSHRIDEDGENTQIEDTEPMSPVLNSKFVPAENDSILMNPAQDGEVQLSQNDDKTKGDDTDTRDDISILATGCKGREETVAEDVCIDLTCDSGSQAVPSPATRSEALSSVLDQEEAMEIKEHHPEEGSSGSEVEEIPETPCESQGEELKEENMESVPLHLSLTETQSQGLCLQKEMPKKECSEAMEVETSVISIDSPQKLAILDQELEHKEQEAWEEATSEDSSVVIVDVKEPSPRVDVSCEPLEGVEKCSDSQSWEDIAPEIEPCAENRLDTKEEKSVEYEGDLKSGTAETEPVEQDSSQPSLPLVRADDPLRLDQELQQPQTQEKTSNSLTEDSKMANAKQLSSDAEAQKLGKPSAHASQSFCESSSETPFHFTLPKEGDIIPPLTGATPPLIGHLKLEPKRHSTPIGISNYPESTIATSDVMSESMVETHDPILGSGKGDSGAAPDVDDKLCLRMKLVSPETEASEESLQFNLEKPATGERKNGSTAVAESVASPQKTMSVLSCICEARQENEARSEDPPTTPIRGNLLHFPSSQGEEEKEKLEGDHTIRQSQQPMKPISPVKDPVSPASQKMVIQGPSSPQGEAMVTDVLEDQKEGRSTNKENPSKALIERPSQNNIGIQTMECSLRVPETVSAATQTIKNVCEQGTSTVDQNFGKQDATVQTERGSGEKPVSAPGDDTESLHSQGEEEFDMPQPPHGHVLHRHMRTIREVRTLVTRVITDVYYVDGTEVERKVTEETEEPIVECQECETEVSPSQTGGSSGDLGDISSFSSKASSLHRTSSGTSLSAMHSSGSSGKGAGPLRGKTSGTEPADFALPSSRGGPGKLSPRKGVSQTGTPVCEEDGDAGLGIRQGGKAPVTPRGRGRRGRPPSRTTGTRETAVPGPLGIEDISPNLSPDDKSFSRVVPRVPDSTRRTDVGAGALRRSDSPEIPFQAAAGPSDGLDASSPGNSFVGLRVVAKWSSNGYFYSGKITRDVGAGKYKLLFDDGYECDVLGKDILLCDPIPLDTEVTALSEDEYFSAGVVKGHRKESGELYYSIEKEGQRKWYKRMAVILSLEQGNRLREQYGLGPYEAVTPLTKAADISLDNLVEGKRKRRSNVSSPATPTASSSSSTTPTRKITESPRASMGVLSGKRKLITSEEERSPAKRGRKSATVKPGAVGAGEFVSPCESGDNTGEPSALEEQRGPLPLNKTLFLGYAFLLTMATTSDKLASRSKLPDGPTGSSEEEEEFLEIPPFNKQYTESQLRAGAGYILEDFNEAQCNTAYQCLLIADQHCRTRKYFLCLASGIPCVSHVWVHDSCHANQLQNYRNYLLPAGYSLEEQRILDWQPRENPFQNLKVLLVSDQQQNFLELWSEILMTGGAASVKQHHSSAHNKDIALGVFDVVVTDPSCPASVLKCAEALQLPVVSQEWVIQCLIVGERIGFKQHPKYKHDYVSH
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Double-strand break (DSB) repair protein involved in response to DNA damage, telomere dynamics and class-switch recombination (CSR) during antibody genesis. Plays a key role in the repair of double-strand DNA breaks (DSBs) in response to DNA damage by promoting non-homologous end joining (NHEJ)-mediated repair of DSBs and specifically counteracting the function of the homologous recombination (HR) repair protein BRCA1. In response to DSBs, phosphorylation by ATM promotes interaction with RIF1 and dissociation from NUDT16L1/TIRR, leading to recruitment to DSBs sites. Recruited to DSBs sites by recognizing and binding histone H2A monoubiquitinated at 'Lys-15' (H2AK15Ub) and histone H4 dimethylated at 'Lys-20' (H4K20me2), two histone marks that are present at DSBs sites. Required for immunoglobulin class-switch recombination (CSR) during antibody genesis, a process that involves the generation of DNA DSBs. Participates to the repair and the orientation of the broken DNA ends during CSR (By similarity). In contrast, it is not required for classic NHEJ and V(D)J recombination (By similarity). Promotes NHEJ of dysfunctional telomeres via interaction with PAXIP1.
Asymmetrically dimethylated on Arg residues by PRMT1. Methylation is required for DNA binding.
Phosphorylated at basal level in the absence of DNA damage. Phosphorylated by ATM in response to DNA damage: phosphorylation at different sites promotes interaction with different set of proteins: phosphorylation at the N-terminus by ATM (residues from 6-178) promotes interaction with PAXIP1 and non-homologous end joining (NHEJ) of dysfunctional telomeres. Phosphorylation by ATM at residues that are located more C-terminus (residues 300-650) leads to promote interaction with RIF1. Interaction with RIF1 leads to disrupt interaction with NUDT16L1/TIRR. Phosphorylation at Thr-1609 and Ser-1618 in the UDR motif blocks interaction with H2AK15ub. Dephosphorylated by PPP4C. Hyperphosphorylation during mitosis correlates with its exclusion from chromatin and DNA lesions. Hyperphosphorylated in an ATR-dependent manner in response to DNA damage induced by UV irradiation. Dephosphorylated by PPP5C.
Nucleus. Chromosome. Chromosome>Centromere>Kinetochore.
Note: Localizes to the nucleus in absence of DNA damage (PubMed:28241136). Following DNA damage, recruited to sites of DNA damage, such as double stand breaks (DSBs): recognizes and binds histone H2A monoubiquitinated at 'Lys-15' (H2AK15Ub) and histone H4 dimethylated at 'Lys-20' (H4K20me2), two histone marks that are present at DSBs sites (PubMed:23333306, PubMed:23760478, PubMed:24703952, PubMed:28241136, PubMed:17190600). Associated with kinetochores during mitosis (By similarity).
The Tudor-like region mediates binding to histone H4 dimethylated at 'Lys-20' (H4K20me2) (PubMed:17190600). Interaction with NUDT16L1/TIRR masks the Tudor-like domain and prevents recruitment to chromatin (PubMed:28241136).
The UDR (ubiquitin-dependent recruitment) motif specifically recognizes and binds histone H2A monoubiquitinated at 'Lys-15' (H2AK15ub) (PubMed:23760478, PubMed:24703952). Phosphorylation of the UDR blocks interaction with H2AK15ub (PubMed:24703952).
Research Fields
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.