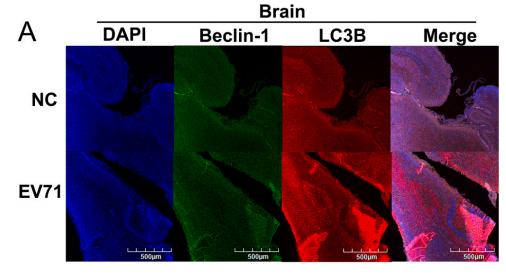
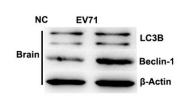
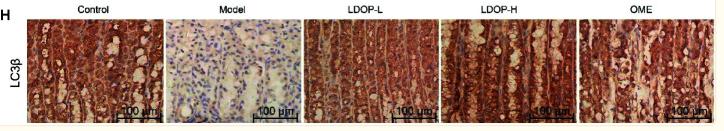
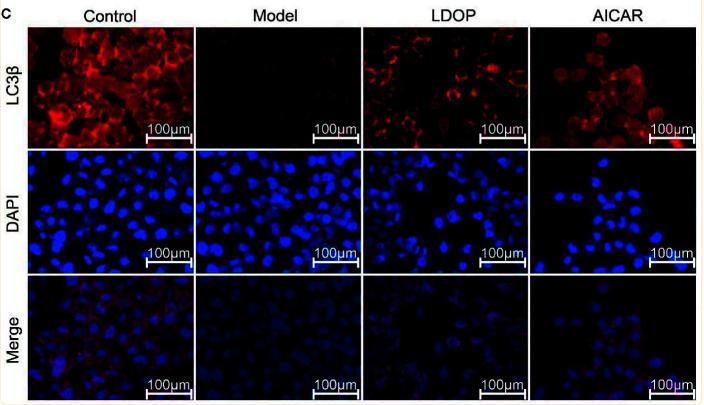
LC3B Antibody - #AF4650
| Product: | LC3B Antibody |
| Catalog: | AF4650 |
| Description: | Rabbit polyclonal antibody to LC3B |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Sheep, Dog, Xenopus |
| Mol.Wt.: | 14kDa,16kDa; 15kD(Calculated). |
| Uniprot: | Q9GZQ8 |
| RRID: | AB_2844592 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF4650, RRID:AB_2844592.
Fold/Unfold
ATG8F; Autophagy-related protein LC3 B; Autophagy-related ubiquitin-like modifier LC3 B; LC3B; LC3II; MAP1 light chain 3 like protein 2; MAP1 light chain 3-like protein 2; MAP1A/1BLC3; MAP1A/MAP1B LC3 B; MAP1A/MAP1B light chain 3 B; MAP1ALC3; MAP1LC3B a; Map1lc3b; Microtubule associated protein 1 light chain 3 beta; Microtubule-associated protein 1 light chain 3 beta; Microtubule-associated proteins 1A/1B light chain 3B; MLP3B_HUMAN;
Immunogens
A synthesized peptide derived from human LC3B, corresponding to a region within N-terminal amino acids.
Most abundant in heart, brain, skeletal muscle and testis. Little expression observed in liver.
- Q9GZQ8 MLP3B_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPSEKTFKQRRTFEQRVEDVRLIREQHPTKIPVIIERYKGEKQLPVLDKTKFLVPDHVNMSELIKIIRRRLQLNANQAFFLLVNGHSMVSVSTPISEVYESEKDEDGFLYMVYASQETFGMKLSV
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Ubiquitin-like modifier involved in formation of autophagosomal vacuoles (autophagosomes). Plays a role in mitophagy which contributes to regulate mitochondrial quantity and quality by eliminating the mitochondria to a basal level to fulfill cellular energy requirements and preventing excess ROS production. Whereas LC3s are involved in elongation of the phagophore membrane, the GABARAP/GATE-16 subfamily is essential for a later stage in autophagosome maturation. Promotes primary ciliogenesis by removing OFD1 from centriolar satellites via the autophagic pathway. Through its interaction with the reticulophagy receptor TEX264, paticipates in the remodeling of subdomains of the endoplasmic reticulum into autophagosomes upon nutrient stress, which then fuse with lysosomes for endoplasmic reticulum turnover.
The precursor molecule is cleaved by ATG4B to form the cytosolic form, LC3-I. This is activated by APG7L/ATG7, transferred to ATG3 and conjugated to phospholipid to form the membrane-bound form, LC3-II.
The Legionella effector RavZ is a deconjugating enzyme that produces an ATG8 product that would be resistant to reconjugation by the host machinery due to the cleavage of the reactive C-terminal glycine.
Phosphorylation at Thr-12 by PKA inhibits conjugation to phosphatidylethanolamine (PE) (By similarity). Interaction with MAPK15 reduces the inhibitory phosphorylation and increases autophagy activity.
Cytoplasm>Cytoskeleton. Endomembrane system>Lipid-anchor. Cytoplasmic vesicle>Autophagosome membrane>Lipid-anchor. Cytoplasmic vesicle>Autophagosome.
Note: LC3-II binds to the autophagic membranes. Localizes also to discrete punctae along the ciliary axoneme (By similarity).
Most abundant in heart, brain, skeletal muscle and testis. Little expression observed in liver.
Belongs to the ATG8 family.
Research Fields
· Cellular Processes > Cell growth and death > Ferroptosis. (View pathway)
References
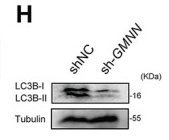
Application: WB Species: mice Sample: bone marrow mesenchymal stem (BMSCs)
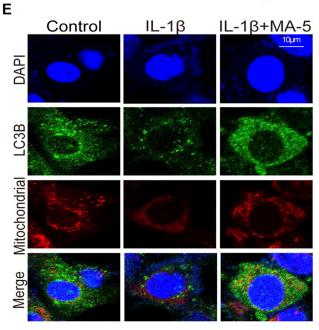
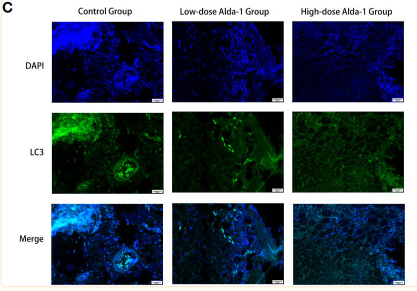
Application: IF/ICC Species: Mouse Sample:
Application: IF/ICC Species: Human Sample: OA cells
Application: WB Species: Human Sample: A2780 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.