RIPK1 Antibody - #AF7877
| Product: | RIPK1 Antibody |
| Catalog: | AF7877 |
| Description: | Rabbit polyclonal antibody to RIPK1 |
| Application: | WB |
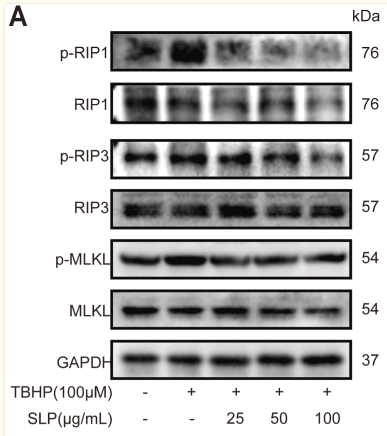
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 76kDa; 76kD(Calculated). |
| Uniprot: | Q13546 |
| RRID: | AB_2844241 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7877, RRID:AB_2844241.
Fold/Unfold
Cell death protein RIP; FLJ39204; OTTHUMP00000039163; Receptor (TNFRSF) interacting serine threonine kinase 1; receptor interacting protein 1; Receptor interacting protein; Receptor interacting protein kinase 1; Receptor interacting serine threonine protein kinase 1; Receptor TNFRSF interacting serine threonine kinase 1; Receptor-interacting protein 1; Receptor-interacting serine/threonine-protein kinase 1; Rinp; RIP 1; RIP; Rip-1; RIP1; RIPK 1; Ripk1; RIPK1_HUMAN; Serine threonine protein kinase RIP; Serine/threonine-protein kinase RIP;
Immunogens
A synthesized peptide derived from human RIPK1, corresponding to a region within the internal amino acids.
- Q13546 RIPK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQPDMSLNVIKMKSSDFLESAELDSGGFGKVSLCFHRTQGLMIMKTVYKGPNCIEHNEALLEEAKMMNRLRHSRVVKLLGVIIEEGKYSLVMEYMEKGNLMHVLKAEMSTPLSVKGRIILEIIEGMCYLHGKGVIHKDLKPENILVDNDFHIKIADLGLASFKMWSKLNNEEHNELREVDGTAKKNGGTLYYMAPEHLNDVNAKPTEKSDVYSFAVVLWAIFANKEPYENAICEQQLIMCIKSGNRPDVDDITEYCPREIISLMKLCWEANPEARPTFPGIEEKFRPFYLSQLEESVEEDVKSLKKEYSNENAVVKRMQSLQLDCVAVPSSRSNSATEQPGSLHSSQGLGMGPVEESWFAPSLEHPQEENEPSLQSKLQDEANYHLYGSRMDRQTKQQPRQNVAYNREEERRRRVSHDPFAQQRPYENFQNTEGKGTAYSSAASHGNAVHQPSGLTSQPQVLYQNNGLYSSHGFGTRPLDPGTAGPRVWYRPIPSHMPSLHNIPVPETNYLGNTPTMPFSSLPPTDESIKYTIYNSTGIQIGAYNYMEIGGTSSSLLDSTNTNFKEEPAAKYQAIFDNTTSLTDKHLDPIRENLGKHWKNCARKLGFTQSQIDEIDHDYERDGLKEKVYQMLQKWVMREGIKGATVGKLAQALHQCSRIDLLSSLIYVSQN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine-threonine kinase which is a key regulator of both cell death and cell survival. Exhibits kinase activity-dependent functions that trigger cell death and kinase-independent scaffold functions regulating inflammatory signaling and cell survival. Initiates ripoptocide which describes cell death that is dependent on RIPK1, be it apoptosis or necroptosis. Upon binding of TNF to TNFR1, RIPK1 is recruited to the TNF-R1 signaling complex (TNF-RSC also known as complex I) where it acts as a scaffold protein promoting cell survival, in part, by activating the canonical NF-kB pathway (By similarity). Specific conditions can however activate RIPK1, and its kinase activity then regulates assembly of two death-inducing complexes, namely complex IIa (RIPK1-FADD-CASP8) and the complex IIb (RIPK1-RIPK3-MLKL) and these complexes respectively drive apoptosis or necroptosis, a regulated form of necrosis. During embryonic development suppresses apoptosis and necroptosis and prevents the interaction of TRADD with FADD thereby limiting aberrant activation of CASP8 (By similarity). Phosphorylates DAB2IP at 'Ser-728' in a TNF- alpha-dependent manner, and thereby activates the MAP3K5-JNK apoptotic cascade. Required for ZBP1-induced NF-kappaB activation and activation of NF-kappaB by DNA damage and IR (By similarity).
Proteolytically cleaved by CASP8 at Asp-324. Cleavage is crucial for limiting apoptosis and necroptosis during embryonic development (By similarity). Cleavage abolishes NF-kappa-B activation and enhances the interaction of TRADD with FADD.
RIPK1 and RIPK3 undergo reciprocal auto- and trans-phosphorylation. Phosphorylation of Ser-161 by RIPK3 is necessary for the formation of the necroptosis-inducing complex. Phosphorylation at Ser-25 represses its kinase activity and consequently prevents TNF-mediated RIPK1-dependent cell death. Phosphorylated at Ser-320 by MAP3K7 which requires prior ubiquitination with 'Lys-63'-linked chains by BIRC2/c-IAP1 and BIRC3/c-IAP2 (By similarity). This phosphorylation positively regulates RIPK1 interaction with RIPK3 to promote necroptosis but negatively regulates RIPK1 kinase activity and its interaction with FADD to mediate apoptosis (By similarity).
Ubiquitinated with 'Lys-11'-, 'Lys-48'-, 'Lys-63'- and linear-linked type ubiquitin Ref.33). Polyubiquitination with 'Lys-63'-linked chains by TRAF2 induces association with the IKK complex. Deubiquitination of 'Lys-63'-linked chains and polyubiquitination with 'Lys-48'-linked chains by TNFAIP3 leads to RIPK1 proteasomal degradation and consequently down-regulates TNF-alpha-induced NFkappa-B signaling. 'Lys-48'-linked polyubiquitination by RFFL or RNF34 also promotes proteasomal degradation and negatively regulates TNF-alpha-induced NF-kappa-B signaling Ref.33). Linear polyubiquitinated; the head-to-tail linear polyubiquitination ('Met-1'-linked) is mediated by the LUBAC complex and decreases protein kinase activity. Deubiquitination of linear polyubiquitin by CYLD promotes the kinase activity (By similarity). Polyubiquitinated with 'Lys-48' and 'Lys-63'-linked chains by BIRC2/c-IAP1 and BIRC3/c-IAP2, leading to activation of NF-kappa-B. Ubiquitinated with 'Lys-63'-linked chains by PELI1. Ubiquitination at Lys-377 with 'Lys-63'-linked chains by BIRC2/c-IAP1 and BIRC3/c-IAP2 is essential for its phosphorylation at Ser-320 mediated by MAP3K7 (By similarity). This ubiquitination is required for NF-kB activation, suppresses RIPK1 kinase activity and plays a critical role in preventing cell death during embryonic development (By similarity).
Cytoplasm. Cell membrane.
The death domain mediates dimerization and activation of its kinase activity during necroptosis and apoptosis (PubMed:29440439). It engages other DD-containing proteins as well as a central (intermediate) region important for NF-kB activation and RHIM-dependent signaling (PubMed:10356400).
Belongs to the protein kinase superfamily. TKL Ser/Thr protein kinase family.
Research Fields
· Cellular Processes > Cell growth and death > Apoptosis. (View pathway)
· Cellular Processes > Cell growth and death > Necroptosis. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Hepatitis C.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Cytosolic DNA-sensing pathway. (View pathway)
References
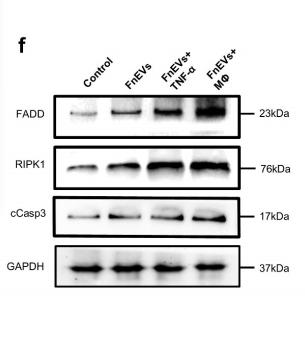
Application: WB Species: human Sample: Caco2 cells
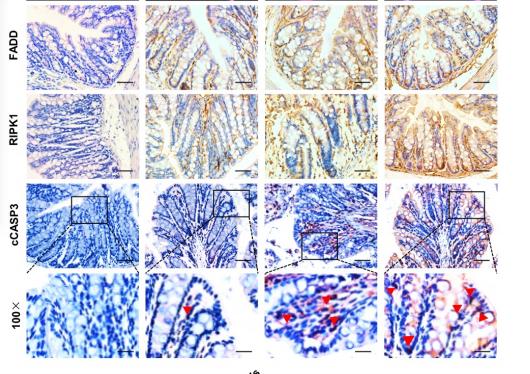
Application: IHC Species: mouse Sample: colon
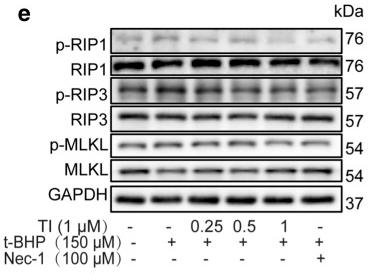
Application: WB Species: Rat Sample: H9c2 cells
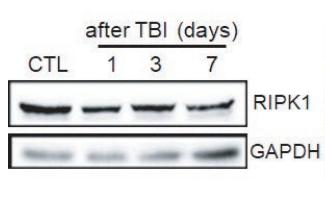
Application: WB Species: mouse Sample: Liver
Application: WB Species: Rat Sample: H9c2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.