p14 ARF Antibody - #AF0229
| Product: | p14 ARF Antibody |
| Catalog: | AF0229 |
| Description: | Rabbit polyclonal antibody to p14 ARF |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human |
| Prediction: | Bovine, Horse, Rabbit, Dog |
| Mol.Wt.: | 18kDa; 14kD(Calculated). |
| Uniprot: | Q8N726 |
| RRID: | AB_2833404 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0229, RRID:AB_2833404.
Fold/Unfold
ARF; CDK4 inhibitor p16 INK4; CDK4I; CDKN2; CDKN2A; Cell cycle negative regulator beta; CMM2; Cyclin dependent kinase 4 inhibitor A; Cyclin dependent kinase inhibitor 2A; INK4; INK4a; Melanoma p16 inhibits CDK4; MLM; MTS 1; MTS1; Multiple tumor suppressor 1; p14; p16; p16 INK4a; p16INK4a; p19; P19ARF; TP16;
Immunogens
A synthesized peptide derived from human p14 ARF, corresponding to a region within the internal amino acids.
- Q8N726 ARF_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MVRRFLVTLRIRRACGPPRVRVFVVHIPRLTGEWAAPGAPAAVALVLMLLRSQRLGQQPLPRRPGHDDGQRPSGGAAAAPRRGAQLRRPRHSHPTRARRCPGGLPGHAGGAAPGRGAAGRARCLGPSARGPG
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Capable of inducing cell cycle arrest in G1 and G2 phases. Acts as a tumor suppressor. Binds to MDM2 and blocks its nucleocytoplasmic shuttling by sequestering it in the nucleolus. This inhibits the oncogenic action of MDM2 by blocking MDM2-induced degradation of p53 and enhancing p53-dependent transactivation and apoptosis. Also induces G2 arrest and apoptosis in a p53-independent manner by preventing the activation of cyclin B1/CDC2 complexes. Binds to BCL6 and down-regulates BCL6-induced transcriptional repression. Binds to E2F1 and MYC and blocks their transcriptional activator activity but has no effect on MYC transcriptional repression. Binds to TOP1/TOPOI and stimulates its activity. This complex binds to rRNA gene promoters and may play a role in rRNA transcription and/or maturation. Interacts with NPM1/B23 and promotes its polyubiquitination and degradation, thus inhibiting rRNA processing. Interacts with COMMD1 and promotes its 'Lys63'-linked polyubiquitination. Interacts with UBE2I/UBC9 and enhances sumoylation of a number of its binding partners including MDM2 and E2F1. Binds to HUWE1 and represses its ubiquitin ligase activity. May play a role in controlling cell proliferation and apoptosis during mammary gland development. Isoform smARF may be involved in regulation of autophagy and caspase-independent cell death; the short-lived mitochondrial isoform is stabilized by C1QBP.
Ubiquitinated in normal cells by TRIP12 via the ubiquitin fusion degradation (UFD) pathway, a process that mediates ubiquitination at the N-terminus, regardless of the absence of lysine residues. Ubiquitination leads to its proteasomal degradation. In cancer cells, however, TRIP12 is located in a different cell compartment, preventing ubiquitination and degradation.
Nucleus>Nucleolus. Nucleus>Nucleoplasm.
Mitochondrion.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Human Diseases > Drug resistance: Antineoplastic > Endocrine resistance.
· Human Diseases > Drug resistance: Antineoplastic > Platinum drug resistance.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Glioma. (View pathway)
· Human Diseases > Cancers: Specific types > Melanoma. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Non-small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
References
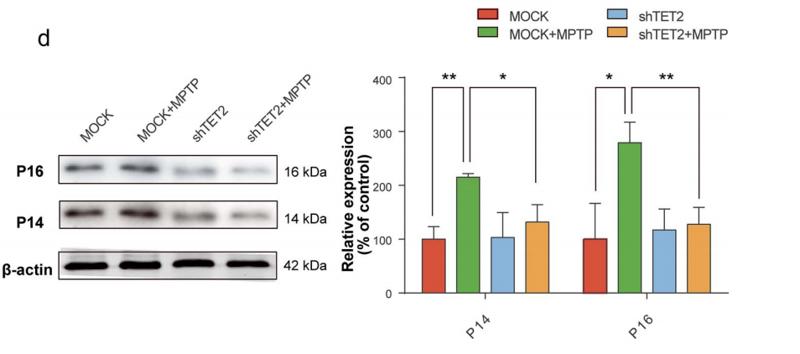
Application: WB Species: Human Sample: hAD-MSCs
Application: WB Species: human Sample: SH-SY5Y cell
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.