Rac1/cdc42 Antibody - #AF7828
| Product: | Rac1/cdc42 Antibody |
| Catalog: | AF7828 |
| Description: | Rabbit polyclonal antibody to Rac1/cdc42 |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 28kDa, 17 kDa; 21kD(Calculated). |
| Uniprot: | P63000 | P60953 |
| RRID: | AB_2844192 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7828, RRID:AB_2844192.
Fold/Unfold
CDC42; CDC42_HUMAN; CDC42Hs; Cell division control protein 42 homolog; Cell division cycle 42 (GTP binding protein 25kDa); Cell division cycle 42; dJ224A6.1.1 (cell division cycle 42 (GTP-binding protein, 25kD)); dJ224A6.1.2 (cell division cycle 42 (GTP-binding protein, 25kD)); G25K; G25K GTP-binding protein; Growth regulating protein; GTP binding protein 25kDa; Small GTP binding protein CDC42; TKS;
Immunogens
A synthesized peptide derived from human Rac1/cdc42, corresponding to a region within the internal amino acids.
Isoform B is predominantly identified in skin and epithelial tissues from the intestinal tract. Its expression is elevated in colorectal tumors at various stages of neoplastic progression, as compared to their respective adjacent tissues.
- P63000 RAC1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQAIKCVVVGDGAVGKTCLLISYTTNAFPGEYIPTVFDNYSANVMVDGKPVNLGLWDTAGQEDYDRLRPLSYPQTDVFLICFSLVSPASFENVRAKWYPEVRHHCPNTPIILVGTKLDLRDDKDTIEKLKEKKLTPITYPQGLAMAKEIGAVKYLECSALTQRGLKTVFDEAIRAVLCPPPVKKRKRKCLLL
- P60953 CDC42_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQTIKCVVVGDGAVGKTCLLISYTTNKFPSEYVPTVFDNYAVTVMIGGEPYTLGLFDTAGQEDYDRLRPLSYPQTDVFLVCFSVVSPSSFENVKEKWVPEITHHCPKTPFLLVGTQIDLRDDPSTIEKLAKNKQKPITPETAEKLARDLKAVKYVECSALTQKGLKNVFDEAILAALEPPEPKKSRRCVLL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Plasma membrane-associated small GTPase which cycles between active GTP-bound and inactive GDP-bound states. In its active state, binds to a variety of effector proteins to regulate cellular responses such as secretory processes, phagocytosis of apoptotic cells, epithelial cell polarization, neurons adhesion, migration and differentiation, and growth-factor induced formation of membrane ruffles. Rac1 p21/rho GDI heterodimer is the active component of the cytosolic factor sigma 1, which is involved in stimulation of the NADPH oxidase activity in macrophages. Essential for the SPATA13-mediated regulation of cell migration and adhesion assembly and disassembly. Stimulates PKN2 kinase activity. In concert with RAB7A, plays a role in regulating the formation of RBs (ruffled borders) in osteoclasts. In podocytes, promotes nuclear shuttling of NR3C2; this modulation is required for a proper kidney functioning. Required for atypical chemokine receptor ACKR2-induced LIMK1-PAK1-dependent phosphorylation of cofilin (CFL1) and for up-regulation of ACKR2 from endosomal compartment to cell membrane, increasing its efficiency in chemokine uptake and degradation. In neurons, is involved in dendritic spine formation and synaptic plasticity (By similarity). In synapses, seems to mediate the regulation of F-actin cluster formation performed by SHANK3.
Isoform B has an accelerated GEF-independent GDP/GTP exchange and an impaired GTP hydrolysis, which is restored partially by GTPase-activating proteins. It is able to bind to the GTPase-binding domain of PAK but not full-length PAK in a GTP-dependent manner, suggesting that the insertion does not completely abolish effector interaction.
(Microbial infection) AMPylation at Tyr-32 and Thr-35 are mediated by bacterial enzymes in case of infection by H.somnus and V.parahaemolyticus, respectively. AMPylation occurs in the effector region and leads to inactivation of the GTPase activity by preventing the interaction with downstream effectors, thereby inhibiting actin assembly in infected cells. It is unclear whether some human enzyme mediates AMPylation; FICD has such ability in vitro but additional experiments remain to be done to confirm results in vivo.
GTP-bound active form is ubiquitinated by HACE1, leading to its degradation by the proteasome.
(Microbial infection) Glycosylated at Tyr-32 by Photorhabdus asymbiotica toxin PAU_02230. Mono-O-GlcNAcylation by PAU_02230 inhibits downstream signaling by an impaired interaction with diverse regulator and effector proteins of Rac and leads to actin disassembly.
Cell membrane>Lipid-anchor>Cytoplasmic side. Melanosome. Cytoplasm. Cell projection>Lamellipodium.
Note: Inner surface of plasma membrane possibly with attachment requiring prenylation of the C-terminal cysteine (PubMed:1903399). Identified by mass spectrometry in melanosome fractions from stage I to stage IV (PubMed:17081065). Found in the ruffled border (a late endosomal-like compartment in the plasma membrane) of bone-resorbing osteoclasts. Localizes to the lamellipodium in a SH3RF1-dependent manner (By similarity). In macrophages, cytoplasmic location increases upon CSF1 stimulation (By similarity).
Isoform B is predominantly identified in skin and epithelial tissues from the intestinal tract. Its expression is elevated in colorectal tumors at various stages of neoplastic progression, as compared to their respective adjacent tissues.
The effector region mediates interaction with DEF6.
Belongs to the small GTPase superfamily. Rho family.
Plasma membrane-associated small GTPase which cycles between an active GTP-bound and an inactive GDP-bound state. In active state binds to a variety of effector proteins to regulate cellular responses. Involved in epithelial cell polarization processes. Regulates the bipolar attachment of spindle microtubules to kinetochores before chromosome congression in metaphase. Regulates cell migration. In neurons, plays a role in the extension and maintenance of the formation of filopodia, thin and actin-rich surface projections. Required for DOCK10-mediated spine formation in Purkinje cells and hippocampal neurons. Facilitates filopodia formation upon DOCK11-activation (By similarity). Upon activation by CaMKII, modulates dendritic spine structural plasticity by relaying CaMKII transient activation to synapse-specific, long-term signaling (By similarity). Also plays a role in phagocytosis through organization of the F-actin cytoskeleton associated with forming phagocytic cups.
(Microbial infection) AMPylation at Tyr-32 and Thr-35 are mediated by bacterial enzymes in case of infection by H.somnus and V.parahaemolyticus, respectively. AMPylation occurs in the effector region and leads to inactivation of the GTPase activity by preventing the interaction with downstream effectors, thereby inhibiting actin assembly in infected cells. It is unclear whether some human enzyme mediates AMPylation; FICD has such ability in vitro but additional experiments remain to be done to confirm results in vivo.
Phosphorylated by SRC in an EGF-dependent manner, this stimulates the binding of the Rho-GDP dissociation inhibitor RhoGDI.
(Microbial infection) Glycosylated at Tyr-32 by Photorhabdus asymbiotica toxin PAU_02230. Mono-O-GlcNAcylation by PAU_02230 inhibits downstream signaling by an impaired interaction with diverse regulator and effector proteins of CDC42 and leads to actin disassembly.
Cell membrane>Lipid-anchor>Cytoplasmic side. Cytoplasm>Cytoskeleton>Microtubule organizing center>Centrosome. Cytoplasm>Cytoskeleton>Spindle. Midbody. Cell projection>Dendrite.
Note: Localizes to spindle during prometaphase cells. Moves to the central spindle as cells progressed through anaphase to telophase (PubMed:15642749). Localizes at the end of cytokinesis in the intercellular bridge formed between two daughter cells (PubMed:15642749). Its localization is regulated by the activities of guanine nucleotide exchange factor ECT2 and GTPase activating protein RACGAP1 (PubMed:15642749). Colocalizes with NEK6 in the centrosome (PubMed:20873783). In its active GTP-bound form localizes to the leading edge membrane of migrating dendritic cells (By similarity).
Belongs to the small GTPase superfamily. Rho family. CDC42 subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Ras signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Non-alcoholic fatty liver disease (NAFLD).
· Human Diseases > Neurodegenerative diseases > Amyotrophic lateral sclerosis (ALS).
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
· Human Diseases > Infectious diseases: Bacterial > Epithelial cell signaling in Helicobacter pylori infection.
· Human Diseases > Infectious diseases: Bacterial > Pathogenic Escherichia coli infection.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Infectious diseases: Bacterial > Salmonella infection.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Pancreatic cancer. (View pathway)
· Human Diseases > Cancers: Overview > Choline metabolism in cancer. (View pathway)
· Human Diseases > Cardiovascular diseases > Viral myocarditis.
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Development > Axon guidance. (View pathway)
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
· Organismal Systems > Immune system > Leukocyte transendothelial migration. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Digestive system > Pancreatic secretion.
References
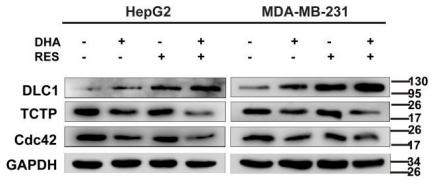
Application: WB Species: Human Sample: HepG2 and MDA-MB-231 cells
Application: WB Species: Human Sample: A2780 and HEY cells
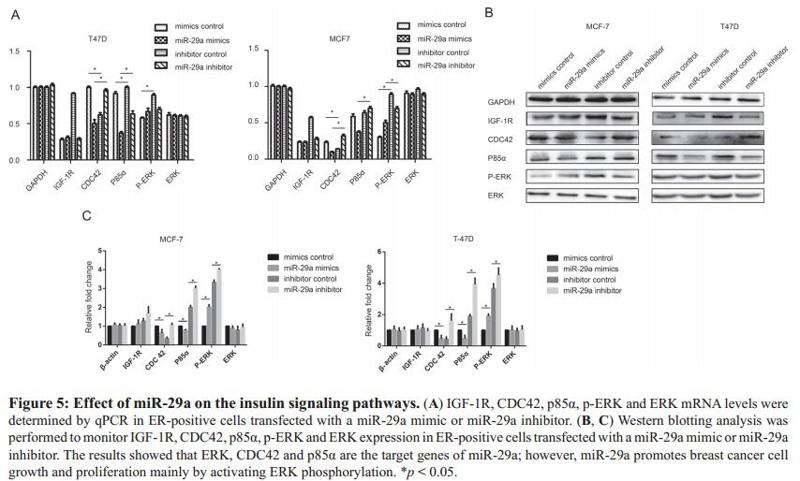
Application: WB Species: human Sample: MCF-7
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.