ATP citrate lyase Antibody - #AF7823
| Product: | ATP citrate lyase Antibody |
| Catalog: | AF7823 |
| Description: | Rabbit polyclonal antibody to ATP citrate lyase |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 125kDa; 121kD(Calculated). |
| Uniprot: | P53396 |
| RRID: | AB_2844187 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7823, RRID:AB_2844187.
Fold/Unfold
ACL; Acly; ACLY_HUMAN; ATP citrate (pro-S) lyase; ATP citrate lyase; ATP citrate synthase; ATP-citrate (pro-S-)-lyase; ATP-citrate synthase; ATPcitrate synthase; ATPCL; Citrate cleavage enzyme; CLATP; OTTHUMP00000164773;
Immunogens
A synthesized peptide derived from human ATP citrate lyase, corresponding to a region within the internal amino acids.
- P53396 ACLY_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSAKAISEQTGKELLYKFICTTSAIQNRFKYARVTPDTDWARLLQDHPWLLSQNLVVKPDQLIKRRGKLGLVGVNLTLDGVKSWLKPRLGQEATVGKATGFLKNFLIEPFVPHSQAEEFYVCIYATREGDYVLFHHEGGVDVGDVDAKAQKLLVGVDEKLNPEDIKKHLLVHAPEDKKEILASFISGLFNFYEDLYFTYLEINPLVVTKDGVYVLDLAAKVDATADYICKVKWGDIEFPPPFGREAYPEEAYIADLDAKSGASLKLTLLNPKGRIWTMVAGGGASVVYSDTICDLGGVNELANYGEYSGAPSEQQTYDYAKTILSLMTREKHPDGKILIIGGSIANFTNVAATFKGIVRAIRDYQGPLKEHEVTIFVRRGGPNYQEGLRVMGEVGKTTGIPIHVFGTETHMTAIVGMALGHRPIPNQPPTAAHTANFLLNASGSTSTPAPSRTASFSESRADEVAPAKKAKPAMPQDSVPSPRSLQGKSTTLFSRHTKAIVWGMQTRAVQGMLDFDYVCSRDEPSVAAMVYPFTGDHKQKFYWGHKEILIPVFKNMADAMRKHPEVDVLINFASLRSAYDSTMETMNYAQIRTIAIIAEGIPEALTRKLIKKADQKGVTIIGPATVGGIKPGCFKIGNTGGMLDNILASKLYRPGSVAYVSRSGGMSNELNNIISRTTDGVYEGVAIGGDRYPGSTFMDHVLRYQDTPGVKMIVVLGEIGGTEEYKICRGIKEGRLTKPIVCWCIGTCATMFSSEVQFGHAGACANQASETAVAKNQALKEAGVFVPRSFDELGEIIQSVYEDLVANGVIVPAQEVPPPTVPMDYSWARELGLIRKPASFMTSICDERGQELIYAGMPITEVFKEEMGIGGVLGLLWFQKRLPKYSCQFIEMCLMVTADHGPAVSGAHNTIICARAGKDLVSSLTSGLLTIGDRFGGALDAAAKMFSKAFDSGIIPMEFVNKMKKEGKLIMGIGHRVKSINNPDMRVQILKDYVRQHFPATPLLDYALEVEKITTSKKPNLILNVDGLIGVAFVDMLRNCGSFTREEADEYIDIGALNGIFVLGRSMGFIGHYLDQKRLKQGLYRHPWDDISYVLPEHMSM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Catalyzes the cleavage of citrate into oxaloacetate and acetyl-CoA, the latter serving as common substrate for de novo cholesterol and fatty acid synthesis.
Phosphorylated by PKA and GSK3 in a sequential manner; phosphorylation results in activation of its activity. Phosphorylation on Thr-447 and Ser-451 depends on the phosphorylation state of Ser-455 (By similarity). Phosphorylation on Ser-455 is decreased by prior phosphorylation on the other 2 residues (By similarity).
ISGylated.
Acetylated at Lys-540, Lys-546 and Lys-554 by KAT2B/PCAF. Acetylation is promoted by glucose and stabilizes the protein, probably by preventing ubiquitination at the same sites. Acetylation promotes de novo lipid synthesis. Deacetylated by SIRT2.
Ubiquitinated at Lys-540, Lys-546 and Lys-554 by UBR4, leading to its degradation. Ubiquitination is probably inhibited by acetylation at same site (Probable).
Cytoplasm>Cytosol.
In the N-terminal section; belongs to the succinate/malate CoA ligase beta subunit family.
In the C-terminal section; belongs to the succinate/malate CoA ligase alpha subunit family.
Research Fields
· Metabolism > Carbohydrate metabolism > Citrate cycle (TCA cycle).
· Metabolism > Global and overview maps > Metabolic pathways.
References
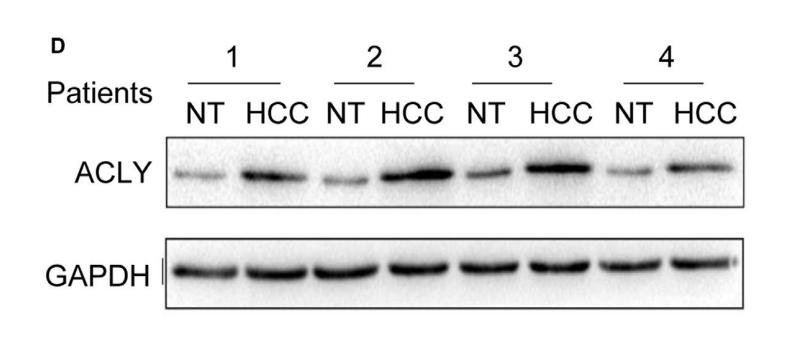
Application: WB Species: Human Sample: liver cancer tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.