CEBP Beta Antibody - #AF7747
| Product: | CEBP Beta Antibody |
| Catalog: | AF7747 |
| Description: | Rabbit polyclonal antibody to CEBP Beta |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Rabbit, Chicken, Xenopus |
| Mol.Wt.: | 36kDa; 36kD(Calculated). |
| Uniprot: | P17676 |
| RRID: | AB_2844111 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7747, RRID:AB_2844111.
Fold/Unfold
AGP/EBP; C EBP beta; C/EBP beta; C/EBP related protein 2; CCAAT Enhancer Binding Protein beta; CCAAT/enhancer-binding protein beta; CEBPB; CEBPB_HUMAN; CRP2; IL 6DBP; IL6DBP; Interleukin 6 dependent binding protein; LAP; Liver activator protein; Liver enriched transcriptional activator; NF IL6; NFIL6; Nuclear factor NF IL6; Nuclear factor NF-IL6; SF B; SFB; Silencer factor B; TCF-5; TCF5; Transcription factor 5;
Immunogens
A synthesized peptide derived from human CEBP Beta, corresponding to a region within the internal amino acids.
- P17676 CEBPB_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQRLVAWDPACLPLPPPPPAFKSMEVANFYYEADCLAAAYGGKAAPAAPPAARPGPRPPAGELGSIGDHERAIDFSPYLEPLGAPQAPAPATATDTFEAAPPAPAPAPASSGQHHDFLSDLFSDDYGGKNCKKPAEYGYVSLGRLGAAKGALHPGCFAPLHPPPPPPPPPAELKAEPGFEPADCKRKEEAGAPGGGAGMAAGFPYALRAYLGYQAVPSGSSGSLSTSSSSSPPGTPSPADAKAPPTACYAGAAPAPSQVKSKAKKTVDKHSDEYKIRRERNNIAVRKSRDKAKMRNLETQHKVLELTAENERLQKKVEQLSRELSTLRNLFKQLPEPLLASSGHC
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Important transcription factor regulating the expression of genes involved in immune and inflammatory responses. Plays also a significant role in adipogenesis, as well as in the gluconeogenic pathway, liver regeneration, and hematopoiesis. The consensus recognition site is 5'-T[TG]NNGNAA[TG]-3'. Its functional capacity is governed by protein interactions and post-translational protein modifications. During early embryogenesis, plays essential and redundant functions with CEBPA. Has a promitotic effect on many cell types such as hepatocytes and adipocytes but has an antiproliferative effect on T-cells by repressing MYC expression, facilitating differentiation along the T-helper 2 lineage. Binds to regulatory regions of several acute-phase and cytokines genes and plays a role in the regulation of acute-phase reaction and inflammation. Plays also a role in intracellular bacteria killing (By similarity). During adipogenesis, is rapidly expressed and, after activation by phosphorylation, induces CEBPA and PPARG, which turn on the series of adipocyte genes that give rise to the adipocyte phenotype. The delayed transactivation of the CEBPA and PPARG genes by CEBPB appears necessary to allow mitotic clonal expansion and thereby progression of terminal differentiation. Essential for female reproduction because of a critical role in ovarian follicle development (By similarity). Restricts osteoclastogenesis: together with NFE2L1; represses expression of DSPP during odontoblast differentiation (By similarity).
Essential for gene expression induction in activated macrophages. Plays a major role in immune responses such as CD4(+) T-cell response, granuloma formation and endotoxin shock. Not essential for intracellular bacteria killing.
Acts as a dominant negative through heterodimerization with isoform 2. Promotes osteoblast differentiation and osteoclastogenesis (By similarity).
Methylated. Methylation at Arg-3 by CARM1 and at Lys-43 by EHMT2 inhibit transactivation activity. Methylation is probably inhibited by phosphorylation at Thr-235.
Sumoylated by polymeric chains of SUMO2 or SUMO3. Sumoylation at Lys-174 is required for inhibition of T-cells proliferation. In adipocytes, sumoylation at Lys-174 by PIAS1 leads to ubiquitination and subsequent proteasomal degradation. Desumoylated by SENP2, which abolishes ubiquitination and stabilizes protein levels (By similarity).
Ubiquitinated, leading to proteasomal degradation.
Phosphorylated at Thr-235 by MAPK and CDK2, serves to prime phosphorylation at Thr-226 and Ser-231 by GSK3B and acquire DNA-binding as well as transactivation activities, required to induce adipogenesis. MAPK and CDK2 act sequentially to maintain Thr-235 in the primed phosphorylated state during mitotical cloning expansion and thereby progression of terminal differentiation. Phosphorylation at Thr-266 enhances transactivation activity. Phosphorylation at Ser-325 in response to calcium increases transactivation activity. Phosphorylated at Thr-235 by RPS6KA1.
O-glycosylated, glycosylation at Ser-227 and Ser-228 prevents phosphorylation on Thr-235, Ser-231 and Thr-226 and DNA binding activity which delays the adipocyte differentiation program.
Acetylated. Acetylation at Lys-43 is an important and dynamic regulatory event that contributes to its ability to transactivate target genes, including those associated with adipogenesis and adipocyte function. Deacetylation by HDAC1 represses its transactivation activity. Acetylated by KAT2A and KAT2B within a cluster of lysine residues between amino acids 129-133, this acetylation is strongly induced by glucocorticoid treatment and enhances transactivation activity.
Nucleus. Cytoplasm.
Note: Translocates to the nucleus when phosphorylated at Ser-288. In T-cells when sumoylated drawn to pericentric heterochromatin thereby allowing proliferation (By similarity).
Expressed at low levels in the lung, kidney and spleen.
the 9aaTAD motif is a transactivation domain present in a large number of yeast and animal transcription factors.
Belongs to the bZIP family. C/EBP subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
References
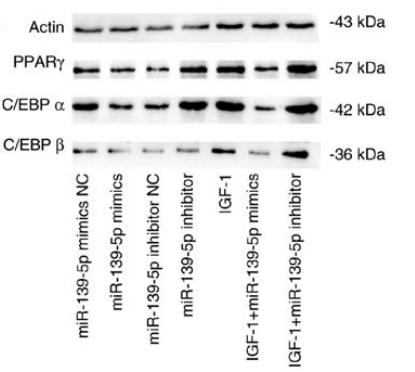
Application: WB Species: human Sample: HemSCs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.