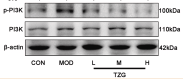
Phospho-PI3 Kinase Class III (Ser244/Ser249) Antibody - #AF7421
| Product: | Phospho-PI3 Kinase Class III (Ser244/Ser249) Antibody |
| Catalog: | AF7421 |
| Description: | Rabbit polyclonal antibody to Phospho-PI3 Kinase Class III (Ser244/Ser249) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Chicken, Xenopus |
| Mol.Wt.: | 100kDa; 102kD(Calculated). |
| Uniprot: | Q8NEB9 |
| RRID: | AB_2843861 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7421, RRID:AB_2843861.
Fold/Unfold
hVps34; MGC61518; Phosphatidylinositol 3 kinase catalytic subunit type 3; Phosphatidylinositol 3 kinase class 3; Phosphatidylinositol 3 kinase p100 subunit; Phosphatidylinositol 3-kinase catalytic subunit type 3; Phosphatidylinositol 3-kinase p100 subunit; Phosphoinositide 3 kinase class 3; Phosphoinositide-3-kinase class 3; PI3 kinase type 3; PI3-kinase type 3; PI3K type 3; Pik3c3; PK3C3_HUMAN; PtdIns 3 kinase type 3; PtdIns-3-kinase type 3; Vps 34; Vps34;
Immunogens
A synthesized peptide derived from human PI3 Kinase Class III around the phosphorylation site of Ser244/249.
- Q8NEB9 PK3C3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGEAEKFHYIYSCDLDINVQLKIGSLEGKREQKSYKAVLEDPMLKFSGLYQETCSDLYVTCQVFAEGKPLALPVRTSYKAFSTRWNWNEWLKLPVKYPDLPRNAQVALTIWDVYGPGKAVPVGGTTVSLFGKYGMFRQGMHDLKVWPNVEADGSEPTKTPGRTSSTLSEDQMSRLAKLTKAHRQGHMVKVDWLDRLTFREIEMINESEKRSSNFMYLMVEFRCVKCDDKEYGIVYYEKDGDESSPILTSFELVKVPDPQMSMENLVESKHHKLARSLRSGPSDHDLKPNAATRDQLNIIVSYPPTKQLTYEEQDLVWKFRYYLTNQEKALTKFLKCVNWDLPQEAKQALELLGKWKPMDVEDSLELLSSHYTNPTVRRYAVARLRQADDEDLLMYLLQLVQALKYENFDDIKNGLEPTKKDSQSSVSENVSNSGINSAEIDSSQIITSPLPSVSSPPPASKTKEVPDGENLEQDLCTFLISRACKNSTLANYLYWYVIVECEDQDTQQRDPKTHEMYLNVMRRFSQALLKGDKSVRVMRSLLAAQQTFVDRLVHLMKAVQRESGNRKKKNERLQALLGDNEKMNLSDVELIPLPLEPQVKIRGIIPETATLFKSALMPAQLFFKTEDGGKYPVIFKHGDDLRQDQLILQIISLMDKLLRKENLDLKLTPYKVLATSTKHGFMQFIQSVPVAEVLDTEGSIQNFFRKYAPSENGPNGISAEVMDTYVKSCAGYCVITYILGVGDRHLDNLLLTKTGKLFHIDFGYILGRDPKPLPPPMKLNKEMVEGMGGTQSEQYQEFRKQCYTAFLHLRRYSNLILNLFSLMVDANIPDIALEPDKTVKKVQDKFRLDLSDEEAVHYMQSLIDESVHALFAAVVEQIHKFAQYWRK
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Catalytic subunit of the PI3K complex that mediates formation of phosphatidylinositol 3-phosphate; different complex forms are believed to play a role in multiple membrane trafficking pathways: PI3KC3-C1 is involved in initiation of autophagosomes and PI3KC3-C2 in maturation of autophagosomes and endocytosis. Involved in regulation of degradative endocytic trafficking and required for the abcission step in cytokinesis, probably in the context of PI3KC3-C2. Involved in the transport of lysosomal enzyme precursors to lysosomes. Required for transport from early to late endosomes (By similarity).
Midbody. Late endosome. Cytoplasmic vesicle>Autophagosome.
Note: As component of the PI3K complex I localized to pre-autophagosome structures. As component of the PI3K complex II localized predominantly to endosomes (Probable). Localizes also to discrete punctae along the ciliary axoneme and to the base of the ciliary axoneme (By similarity).
Ubiquitously expressed, with a highest expression in skeletal muscle.
Belongs to the PI3/PI4-kinase family.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - other. (View pathway)
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
· Environmental Information Processing > Signal transduction > Phosphatidylinositol signaling system.
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Metabolism > Carbohydrate metabolism > Inositol phosphate metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
References
Application: WB Species: Rat Sample:
Application: WB Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.