Phospho-RUNX2 (Ser275) Antibody - #AF7379
| Product: | Phospho-RUNX2 (Ser275) Antibody |
| Catalog: | AF7379 |
| Description: | Rabbit polyclonal antibody to Phospho-RUNX2 (Ser275) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Chicken, Xenopus |
| Mol.Wt.: | 50-60kd; 57kD(Calculated). |
| Uniprot: | Q13950 |
| RRID: | AB_2843819 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7379, RRID:AB_2843819.
Fold/Unfold
Acute myeloid leukemia 3 protein; Alpha subunit 1; AML3; CBF alpha 1; CBF-alpha-1; CBFA1; CCD; CCD1; Cleidocranial dysplasia 1; Core binding factor; Core binding factor runt domain alpha subunit 1; Core binding factor subunit alpha 1; Core-binding factor subunit alpha-1; MGC120022; MGC120023; Oncogene AML 3; Oncogene AML-3; OSF 2; OSF-2; OSF2; Osteoblast specific transcription factor 2; Osteoblast-specific transcription factor 2; OTTHUMP00000016533; PEA2 alpha A; PEA2-alpha A; PEA2aA; PEBP2 alpha A; PEBP2-alpha A; PEBP2A1; PEBP2A2; PEBP2aA; PEBP2aA1; Polyomavirus enhancer binding protein 2 alpha A subunit; Polyomavirus enhancer-binding protein 2 alpha A subunit; Runt domain; Runt related transcription factor 2; Runt-related transcription factor 2; RUNX2; RUNX2_HUMAN; SL3 3 enhancer factor 1 alpha A subunit; SL3-3 enhancer factor 1 alpha A subunit; SL3/AKV core binding factor alpha A subunit; SL3/AKV core-binding factor alpha A subunit;
Immunogens
A synthesized peptide derived from human RUNX2 around the phosphorylation site of Ser275.
- Q13950 RUNX2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MASNSLFSTVTPCQQNFFWDPSTSRRFSPPSSSLQPGKMSDVSPVVAAQQQQQQQQQQQQQQQQQQQQQQQEAAAAAAAAAAAAAAAAAVPRLRPPHDNRTMVEIIADHPAELVRTDSPNFLCSVLPSHWRCNKTLPVAFKVVALGEVPDGTVVTVMAGNDENYSAELRNASAVMKNQVARFNDLRFVGRSGRGKSFTLTITVFTNPPQVATYHRAIKVTVDGPREPRRHRQKLDDSKPSLFSDRLSDLGRIPHPSMRVGVPPQNPRPSLNSAPSPFNPQGQSQITDPRQAQSSPPWSYDQSYPSYLSQMTSPSIHSTTPLSSTRGTGLPAITDVPRRISDDDTATSDFCLWPSTLSKKSQAGASELGPFSDPRQFPSISSLTESRFSNPRMHYPATFTYTPPVTSGMSLGMSATTHYHTYLPPPYPGSSQSQSGPFQTSSTPYLYYGTSSGSYQFPMVPGGDRSPSRMLPPCTTTSNGSTLLNPNLPNQNDGVDADGSHSSSPTVLNSSGRMDESVWRPY
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcription factor involved in osteoblastic differentiation and skeletal morphogenesis. Essential for the maturation of osteoblasts and both intramembranous and endochondral ossification. CBF binds to the core site, 5'-PYGPYGGT-3', of a number of enhancers and promoters, including murine leukemia virus, polyomavirus enhancer, T-cell receptor enhancers, osteocalcin, osteopontin, bone sialoprotein, alpha 1(I) collagen, LCK, IL-3 and GM-CSF promoters. In osteoblasts, supports transcription activation: synergizes with SPEN/MINT to enhance FGFR2-mediated activation of the osteocalcin FGF-responsive element (OCFRE) (By similarity). Inhibits KAT6B-dependent transcriptional activation.
Phosphorylated; probably by MAP kinases (MAPK). Phosphorylation by HIPK3 is required for the SPEN/MINT and FGF2 transactivation during osteoblastic differentiation (By similarity). Phosphorylation at Ser-451 by CDK1 promotes endothelial cell proliferation required for tumor angiogenesis probably by facilitating cell cycle progression. Isoform 3 is phosphorylated on Ser-340.
Nucleus.
Specifically expressed in osteoblasts.
A proline/serine/threonine rich region at the C-terminus is necessary for transcriptional activation of target genes and contains the phosphorylation sites.
Research Fields
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
References
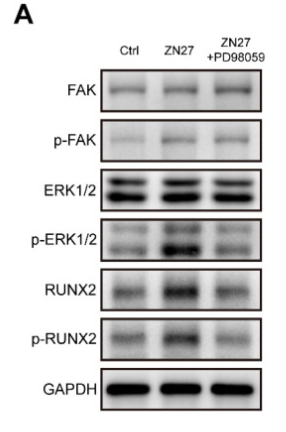
Application: WB Species: Human Sample: trigonocephaly cranial suture cells
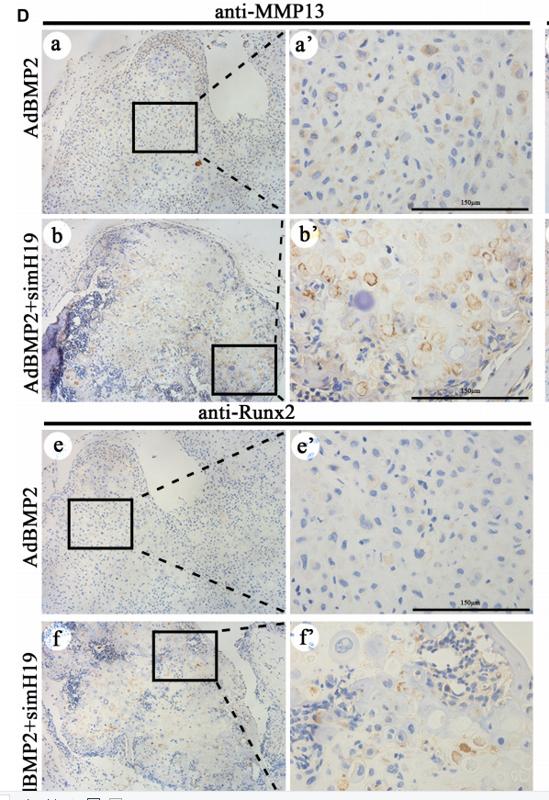
Application: IHC Species: mice Sample: flanks
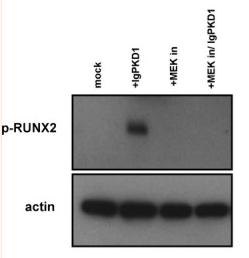
Application: WB Species: Rat Sample:
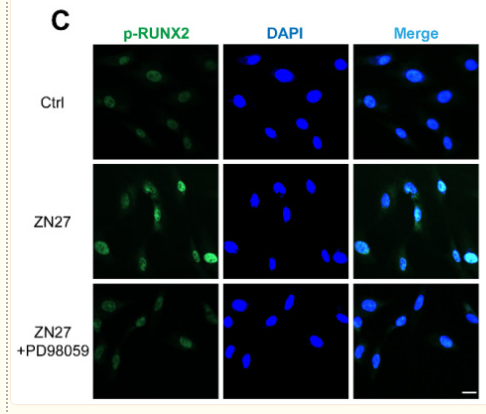
Application: IF/ICC Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.