Phospho-PKC epsilon (Thr710) Antibody - #AF7345
| Product: | Phospho-PKC epsilon (Thr710) Antibody |
| Catalog: | AF7345 |
| Description: | Rabbit polyclonal antibody to Phospho-PKC epsilon (Thr710) |
| Application: | WB |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Sheep, Rabbit, Dog |
| Mol.Wt.: | 84kDa; 84kD(Calculated). |
| Uniprot: | Q02156 |
| RRID: | AB_2843785 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF7345, RRID:AB_2843785.
Fold/Unfold
KPCE_HUMAN; MGC125656; MGC125657; nPKC epsilon; nPKC-epsilon; PKCE; Pkcea; PRKCE; Protein kinase C epsilon; Protein kinase C epsilon type;
Immunogens
A synthesized peptide derived from human PKC around the phosphorylation site of Thr710.
- Q02156 KPCE_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MVVFNGLLKIKICEAVSLKPTAWSLRHAVGPRPQTFLLDPYIALNVDDSRIGQTATKQKTNSPAWHDEFVTDVCNGRKIELAVFHDAPIGYDDFVANCTIQFEELLQNGSRHFEDWIDLEPEGRVYVIIDLSGSSGEAPKDNEERVFRERMRPRKRQGAVRRRVHQVNGHKFMATYLRQPTYCSHCRDFIWGVIGKQGYQCQVCTCVVHKRCHELIITKCAGLKKQETPDQVGSQRFSVNMPHKFGIHNYKVPTFCDHCGSLLWGLLRQGLQCKVCKMNVHRRCETNVAPNCGVDARGIAKVLADLGVTPDKITNSGQRRKKLIAGAESPQPASGSSPSEEDRSKSAPTSPCDQEIKELENNIRKALSFDNRGEEHRAASSPDGQLMSPGENGEVRQGQAKRLGLDEFNFIKVLGKGSFGKVMLAELKGKDEVYAVKVLKKDVILQDDDVDCTMTEKRILALARKHPYLTQLYCCFQTKDRLFFVMEYVNGGDLMFQIQRSRKFDEPRSRFYAAEVTSALMFLHQHGVIYRDLKLDNILLDAEGHCKLADFGMCKEGILNGVTTTTFCGTPDYIAPEILQELEYGPSVDWWALGVLMYEMMAGQPPFEADNEDDLFESILHDDVLYPVWLSKEAVSILKAFMTKNPHKRLGCVASQNGEDAIKQHPFFKEIDWVLLEQKKIKPPFKPRIKTKRDVNNFDQDFTREEPVLTLVDEAIVKQINQEEFKGFSYFGEDLMP
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Calcium-independent, phospholipid- and diacylglycerol (DAG)-dependent serine/threonine-protein kinase that plays essential roles in the regulation of multiple cellular processes linked to cytoskeletal proteins, such as cell adhesion, motility, migration and cell cycle, functions in neuron growth and ion channel regulation, and is involved in immune response, cancer cell invasion and regulation of apoptosis. Mediates cell adhesion to the extracellular matrix via integrin-dependent signaling, by mediating angiotensin-2-induced activation of integrin beta-1 (ITGB1) in cardiac fibroblasts. Phosphorylates MARCKS, which phosphorylates and activates PTK2/FAK, leading to the spread of cardiomyocytes. Involved in the control of the directional transport of ITGB1 in mesenchymal cells by phosphorylating vimentin (VIM), an intermediate filament (IF) protein. In epithelial cells, associates with and phosphorylates keratin-8 (KRT8), which induces targeting of desmoplakin at desmosomes and regulates cell-cell contact. Phosphorylates IQGAP1, which binds to CDC42, mediating epithelial cell-cell detachment prior to migration. In HeLa cells, contributes to hepatocyte growth factor (HGF)-induced cell migration, and in human corneal epithelial cells, plays a critical role in wound healing after activation by HGF. During cytokinesis, forms a complex with YWHAB, which is crucial for daughter cell separation, and facilitates abscission by a mechanism which may implicate the regulation of RHOA. In cardiac myocytes, regulates myofilament function and excitation coupling at the Z-lines, where it is indirectly associated with F-actin via interaction with COPB1. During endothelin-induced cardiomyocyte hypertrophy, mediates activation of PTK2/FAK, which is critical for cardiomyocyte survival and regulation of sarcomere length. Plays a role in the pathogenesis of dilated cardiomyopathy via persistent phosphorylation of troponin I (TNNI3). Involved in nerve growth factor (NFG)-induced neurite outgrowth and neuron morphological change independently of its kinase activity, by inhibition of RHOA pathway, activation of CDC42 and cytoskeletal rearrangement. May be involved in presynaptic facilitation by mediating phorbol ester-induced synaptic potentiation. Phosphorylates gamma-aminobutyric acid receptor subunit gamma-2 (GABRG2), which reduces the response of GABA receptors to ethanol and benzodiazepines and may mediate acute tolerance to the intoxicating effects of ethanol. Upon PMA treatment, phosphorylates the capsaicin- and heat-activated cation channel TRPV1, which is required for bradykinin-induced sensitization of the heat response in nociceptive neurons. Is able to form a complex with PDLIM5 and N-type calcium channel, and may enhance channel activities and potentiates fast synaptic transmission by phosphorylating the pore-forming alpha subunit CACNA1B (CaV2.2). In prostate cancer cells, interacts with and phosphorylates STAT3, which increases DNA-binding and transcriptional activity of STAT3 and seems to be essential for prostate cancer cell invasion. Downstream of TLR4, plays an important role in the lipopolysaccharide (LPS)-induced immune response by phosphorylating and activating TICAM2/TRAM, which in turn activates the transcription factor IRF3 and subsequent cytokines production. In differentiating erythroid progenitors, is regulated by EPO and controls the protection against the TNFSF10/TRAIL-mediated apoptosis, via BCL2. May be involved in the regulation of the insulin-induced phosphorylation and activation of AKT1.
Phosphorylation on Thr-566 by PDPK1 triggers autophosphorylation on Ser-729. Phosphorylation in the hinge domain at Ser-350 by MAPK11 or MAPK14, Ser-346 by GSK3B and Ser-368 by autophosphorylation is required for interaction with YWHAB.
Cytoplasm. Cytoplasm>Cytoskeleton. Cell membrane. Cytoplasm>Perinuclear region. Nucleus.
Note: Translocated to plasma membrane in epithelial cells stimulated by HGF. Associated with the Golgi at the perinuclear site in pre-passage fibroblasts (By similarity). In passaging cells, translocated to the cell periphery (By similarity). Translocated to the nucleus in PMA-treated cells (By similarity).
The C1 domain, containing the phorbol ester/DAG-type region 1 (C1A) and 2 (C1B), is the diacylglycerol sensor and the C2 domain is a non-calcium binding domain.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. PKC subfamily.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Sphingolipid signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Apelin signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Type II diabetes mellitus.
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Organismal Systems > Circulatory system > Vascular smooth muscle contraction. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
· Organismal Systems > Sensory system > Inflammatory mediator regulation of TRP channels. (View pathway)
· Organismal Systems > Endocrine system > Aldosterone synthesis and secretion.
References
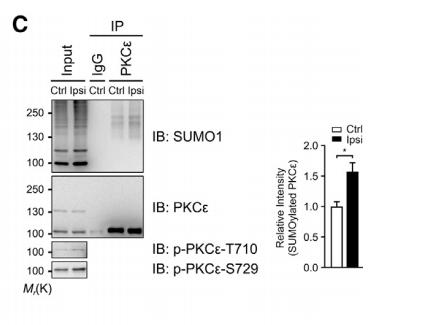
Application: WB Species: mice Sample: hindpaws
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.