MCM2 Antibody - #AF0206
| Product: | MCM2 Antibody |
| Catalog: | AF0206 |
| Description: | Rabbit polyclonal antibody to MCM2 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Horse, Rabbit, Dog |
| Mol.Wt.: | 120kDa; 102kD(Calculated). |
| Uniprot: | P49736 |
| RRID: | AB_2833393 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0206, RRID:AB_2833393.
Fold/Unfold
BM28; CCNL 1; CCNL1; CDC like 1; cdc19; CDCL 1; CDCL1; Cell devision cycle like 1; Cyclin like 1; D3S3194; DNA replication licensing factor MCM2; KIAA0030; MCM 2; MCM2; MCM2 minichromosome maintenance deficient 2 mitotin; MCM2 minichromosome maintenance deficient 2 mitotin (S. cerevisiae); MCM2 minichromosome maintenance deficient 2, mitotin; MCM2_HUMAN; MGC10606; Minichromosome maintenance complex component 2; Minichromosome maintenance deficient 2 (mitotin); Minichromosome maintenance deficient 2 mitotin; Minichromosome maintenance protein 2; Minichromosome maintenance protein 2 homolog; Mitotin; Nuclear protein BM28; OTTHUMP00000216047; OTTHUMP00000216050;
Immunogens
A synthesized peptide derived from human MCM2, corresponding to a region within N-terminal amino acids.
- P49736 MCM2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAESSESFTMASSPAQRRRGNDPLTSSPGRSSRRTDALTSSPGRDLPPFEDESEGLLGTEGPLEEEEDGEELIGDGMERDYRAIPELDAYEAEGLALDDEDVEELTASQREAAERAMRQRDREAGRGLGRMRRGLLYDSDEEDEERPARKRRQVERATEDGEEDEEMIESIENLEDLKGHSVREWVSMAGPRLEIHHRFKNFLRTHVDSHGHNVFKERISDMCKENRESLVVNYEDLAAREHVLAYFLPEAPAELLQIFDEAALEVVLAMYPKYDRITNHIHVRISHLPLVEELRSLRQLHLNQLIRTSGVVTSCTGVLPQLSMVKYNCNKCNFVLGPFCQSQNQEVKPGSCPECQSAGPFEVNMEETIYQNYQRIRIQESPGKVAAGRLPRSKDAILLADLVDSCKPGDEIELTGIYHNNYDGSLNTANGFPVFATVILANHVAKKDNKVAVGELTDEDVKMITSLSKDQQIGEKIFASIAPSIYGHEDIKRGLALALFGGEPKNPGGKHKVRGDINVLLCGDPGTAKSQFLKYIEKVSSRAIFTTGQGASAVGLTAYVQRHPVSREWTLEAGALVLADRGVCLIDEFDKMNDQDRTSIHEAMEQQSISISKAGIVTSLQARCTVIAAANPIGGRYDPSLTFSENVDLTEPIISRFDILCVVRDTVDPVQDEMLARFVVGSHVRHHPSNKEEEGLANGSAAEPAMPNTYGVEPLPQEVLKKYIIYAKERVHPKLNQMDQDKVAKMYSDLRKESMATGSIPITVRHIESMIRMAEAHARIHLRDYVIEDDVNMAIRVMLESFIDTQKFSVMRSMRKTFARYLSFRRDNNELLLFILKQLVAEQVTYQRNRFGAQQDTIEVPEKDLVDKARQINIHNLSAFYDSELFRMNKFSHDLKRKMILQQF
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Acts as component of the MCM2-7 complex (MCM complex) which is the putative replicative helicase essential for 'once per cell cycle' DNA replication initiation and elongation in eukaryotic cells. The active ATPase sites in the MCM2-7 ring are formed through the interaction surfaces of two neighboring subunits such that a critical structure of a conserved arginine finger motif is provided in trans relative to the ATP-binding site of the Walker A box of the adjacent subunit. The six ATPase active sites, however, are likely to contribute differentially to the complex helicase activity. Required for the entry in S phase and for cell division. Plays a role in terminally differentiated hair cells development of the cochlea and induces cells apoptosis.
Phosphorylated on Ser-108 by ATR in proliferating cells. Ser-108 proliferation is increased by genotoxic agents. Ser-40 is mediated by the CDC7-DBF4 and CDC7-DBF4B complexes, while Ser-53 phosphorylation is only mediated by the CDC7-DBF4 complex. Phosphorylation by the CDC7-DBF4 complex during G1/S phase is required for the initiation of DNA replication.
Nucleus.
Belongs to the MCM family.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Genetic Information Processing > Replication and repair > DNA replication.
References
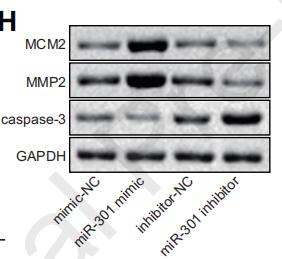
Application: WB Species: Human Sample: breast cancer cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.