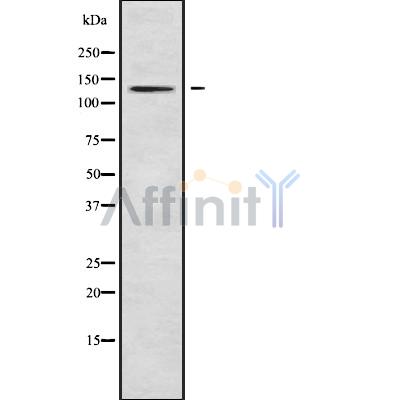
ATP2B1/2/3 Antibody - #DF9758
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF9758, RRID:AB_2842953.
Fold/Unfold
AT2B1_HUMAN; ATP2B1; ATP2B2; ATP2B3; ATP2B4; ATPase Ca++ transporting plasma membrane 1; ATPase Ca++ transporting plasma membrane 2; ATPase Ca++ transporting plasma membrane 3; ATPase Ca++ transporting plasma membrane 4; Plasma membrane calcium ATPase isoform 1; Plasma membrane calcium pump isoform 1; Plasma membrane calcium transporting ATPase 1; Plasma membrane calcium transporting ATPase 2; Plasma membrane calcium transporting ATPase 3; Plasma membrane calcium transporting ATPase 4; Plasma membrane calcium-transporting ATPase 1; PMCA1; PMCA2; PMCA3; PMCA4; AT2B1_HUMAN; ATP2B1; ATPase Ca++ transporting plasma membrane 1; Plasma membrane calcium ATPase 1; Plasma membrane calcium ATPase isoform 1; Plasma membrane calcium pump isoform 1; Plasma membrane calcium transporting ATPase 1; Plasma membrane calcium-transporting ATPase 1; PMCA 1; PMCA1; AT2B2_HUMAN; atp2b2; ATPase plasma membrane Ca2+ transporting 2; Plasma membrane calcium ATPase 2; Plasma membrane calcium ATPase; Plasma membrane calcium ATPase isoform 2; Plasma membrane calcium pump isoform 2; Plasma membrane calcium transporting ATPase 2; Plasma membrane calcium-transporting ATPase 2; PMCA2; PMCA2a; PMCA2i;
Immunogens
A synthesized peptide derived from human ATP2B1/2/3, corresponding to a region within C-terminal amino acids.
Highly expressed in the cerebellum, particulary in the presynaptic terminals of parallel fibers-Purkinje neurons. Isoform XE and isoform XB are the most abundant isoforms and are detected at low levels in brain and fetal skeletal muscle. The other isoforms are only found at lower levels and not in fetal tissues.
P20020 AT2B1_HUMAN:Isoform B: Ubiquitously expressed. Isoform C: Found in brain cortex, skeletal muscle and heart muscle. Isoform D: Has only been found in fetal skeletal muscle. Isoform K: Found in small intestine and liver. Abundantly expressed in the endometrial epithelial cells and glandular epithelial cells in early-proliferative phase and early-secretory phases (PubMed:21400627).
Q01814 AT2B2_HUMAN:Mainly expressed in brain cortex. Found in low levels in skeletal muscle, heart muscle, stomach, liver, kidney and lung. Isoforms containing segment B are found in brain cortex and at low levels in other tissues. Isoforms containing segments X and W are found at low levels in all tissues. Isoforms containing segment A and segment Z are found at low levels in skeletal muscle and heart muscle.
- Q16720 AT2B3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGDMANSSIEFHPKPQQQRDVPQAGGFGCTLAELRTLMELRGAEALQKIEEAYGDVSGLCRRLKTSPTEGLADNTNDLEKRRQIYGQNFIPPKQPKTFLQLVWEALQDVTLIILEVAAIVSLGLSFYAPPGEESEACGNVSGGAEDEGEAEAGWIEGAAILLSVICVVLVTAFNDWSKEKQFRGLQSRIEQEQKFTVIRNGQLLQVPVAALVVGDIAQVKYGDLLPADGVLIQANDLKIDESSLTGESDHVRKSADKDPMLLSGTHVMEGSGRMVVTAVGVNSQTGIIFTLLGAGGEEEEKKDKKGKQQDGAMESSQTKAKKQDGAVAMEMQPLKSAEGGEMEEREKKKANAPKKEKSVLQGKLTKLAVQIGKAGLVMSAITVIILVLYFVIETFVVEGRTWLAECTPVYVQYFVKFFIIGVTVLVVAVPEGLPLAVTISLAYSVKKMMKDNNLVRHLDACETMGNATAICSDKTGTLTTNRMTVVQSYLGDTHYKEIPAPSALTPKILDLLVHAISINSAYTTKILPPEKEGALPRQVGNKTECALLGFVLDLKRDFQPVREQIPEDKLYKVYTFNSVRKSMSTVIRMPDGGFRLFSKGASEILLKKCTNILNSNGELRGFRPRDRDDMVRKIIEPMACDGLRTICIAYRDFSAGQEPDWDNENEVVGDLTCIAVVGIEDPVRPEVPEAIRKCQRAGITVRMVTGDNINTARAIAAKCGIIQPGEDFLCLEGKEFNRRIRNEKGEIEQERLDKVWPKLRVLARSSPTDKHTLVKGIIDSTTGEQRQVVAVTGDGTNDGPALKKADVGFAMGIAGTDVAKEASDIILTDDNFTSIVKAVMWGRNVYDSISKFLQFQLTVNVVAVIVAFTGACITQDSPLKAVQMLWVNLIMDTFASLALATEPPTESLLLRKPYGRDKPLISRTMMKNILGHAVYQLAIIFTLLFVGELFFDIDSGRNAPLHSPPSEHYTIIFNTFVMMQLFNEINARKIHGERNVFDGIFSNPIFCTIVLGTFGIQIVIVQFGGKPFSCSPLSTEQWLWCLFVGVGELVWGQVIATIPTSQLKCLKEAGHGPGKDEMTDEELAEGEEEIDHAERELRRGQILWFRGLNRIQTQIRVVKAFRSSLYEGLEKPESKTSIHNFMATPEFLINDYTHNIPLIDDTDVDENEERLRAPPPPSPNQNNNAIDSGIYLTTHVTKSATSSVFSSSPGSPLHSVETSL
- P20020 AT2B1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGDMANNSVAYSGVKNSLKEANHDGDFGITLAELRALMELRSTDALRKIQESYGDVYGICTKLKTSPNEGLSGNPADLERREAVFGKNFIPPKKPKTFLQLVWEALQDVTLIILEIAAIVSLGLSFYQPPEGDNALCGEVSVGEEEGEGETGWIEGAAILLSVVCVVLVTAFNDWSKEKQFRGLQSRIEQEQKFTVIRGGQVIQIPVADITVGDIAQVKYGDLLPADGILIQGNDLKIDESSLTGESDHVKKSLDKDPLLLSGTHVMEGSGRMVVTAVGVNSQTGIIFTLLGAGGEEEEKKDEKKKEKKNKKQDGAIENRNKAKAQDGAAMEMQPLKSEEGGDGDEKDKKKANLPKKEKSVLQGKLTKLAVQIGKAGLLMSAITVIILVLYFVIDTFWVQKRPWLAECTPIYIQYFVKFFIIGVTVLVVAVPEGLPLAVTISLAYSVKKMMKDNNLVRHLDACETMGNATAICSDKTGTLTMNRMTVVQAYINEKHYKKVPEPEAIPPNILSYLVTGISVNCAYTSKILPPEKEGGLPRHVGNKTECALLGLLLDLKRDYQDVRNEIPEEALYKVYTFNSVRKSMSTVLKNSDGSYRIFSKGASEIILKKCFKILSANGEAKVFRPRDRDDIVKTVIEPMASEGLRTICLAFRDFPAGEPEPEWDNENDIVTGLTCIAVVGIEDPVRPEVPDAIKKCQRAGITVRMVTGDNINTARAIATKCGILHPGEDFLCLEGKDFNRRIRNEKGEIEQERIDKIWPKLRVLARSSPTDKHTLVKGIIDSTVSDQRQVVAVTGDGTNDGPALKKADVGFAMGIAGTDVAKEASDIILTDDNFTSIVKAVMWGRNVYDSISKFLQFQLTVNVVAVIVAFTGACITQDSPLKAVQMLWVNLIMDTLASLALATEPPTESLLLRKPYGRNKPLISRTMMKNILGHAFYQLVVVFTLLFAGEKFFDIDSGRNAPLHAPPSEHYTIVFNTFVLMQLFNEINARKIHGERNVFEGIFNNAIFCTIVLGTFVVQIIIVQFGGKPFSCSELSIEQWLWSIFLGMGTLLWGQLISTIPTSRLKFLKEAGHGTQKEEIPEEELAEDVEEIDHAERELRRGQILWFRGLNRIQTQIRVVNAFRSSLYEGLEKPESRSSIHNFMTHPEFRIEDSEPHIPLIDDTDAEDDAPTKRNSSPPPSPNKNNNAVDSGIHLTIEMNKSATSSSPGSPLHSLETSL
- Q01814 AT2B2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGDMTNSDFYSKNQRNESSHGGEFGCTMEELRSLMELRGTEAVVKIKETYGDTEAICRRLKTSPVEGLPGTAPDLEKRKQIFGQNFIPPKKPKTFLQLVWEALQDVTLIILEIAAIISLGLSFYHPPGEGNEGCATAQGGAEDEGEAEAGWIEGAAILLSVICVVLVTAFNDWSKEKQFRGLQSRIEQEQKFTVVRAGQVVQIPVAEIVVGDIAQVKYGDLLPADGLFIQGNDLKIDESSLTGESDQVRKSVDKDPMLLSGTHVMEGSGRMLVTAVGVNSQTGIIFTLLGAGGEEEEKKDKKGVKKGDGLQLPAADGAAASNAADSANASLVNGKMQDGNVDASQSKAKQQDGAAAMEMQPLKSAEGGDADDRKKASMHKKEKSVLQGKLTKLAVQIGKAGLVMSAITVIILVLYFTVDTFVVNKKPWLPECTPVYVQYFVKFFIIGVTVLVVAVPEGLPLAVTISLAYSVKKMMKDNNLVRHLDACETMGNATAICSDKTGTLTTNRMTVVQAYVGDVHYKEIPDPSSINTKTMELLINAIAINSAYTTKILPPEKEGALPRQVGNKTECGLLGFVLDLKQDYEPVRSQMPEEKLYKVYTFNSVRKSMSTVIKLPDESFRMYSKGASEIVLKKCCKILNGAGEPRVFRPRDRDEMVKKVIEPMACDGLRTICVAYRDFPSSPEPDWDNENDILNELTCICVVGIEDPVRPEVPEAIRKCQRAGITVRMVTGDNINTARAIAIKCGIIHPGEDFLCLEGKEFNRRIRNEKGEIEQERIDKIWPKLRVLARSSPTDKHTLVKGIIDSTHTEQRQVVAVTGDGTNDGPALKKADVGFAMGIAGTDVAKEASDIILTDDNFSSIVKAVMWGRNVYDSISKFLQFQLTVNVVAVIVAFTGACITQDSPLKAVQMLWVNLIMDTFASLALATEPPTETLLLRKPYGRNKPLISRTMMKNILGHAVYQLALIFTLLFVGEKMFQIDSGRNAPLHSPPSEHYTIIFNTFVMMQLFNEINARKIHGERNVFDGIFRNPIFCTIVLGTFAIQIVIVQFGGKPFSCSPLQLDQWMWCIFIGLGELVWGQVIATIPTSRLKFLKEAGRLTQKEEIPEEELNEDVEEIDHAERELRRGQILWFRGLNRIQTQIRVVKAFRSSLYEGLEKPESRTSIHNFMAHPEFRIEDSQPHIPLIDDTDLEEDAALKQNSSPPSSLNKNNSAIDSGINLTTDTSKSATSSSPGSPIHSLETSL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell.
Cell membrane>Multi-pass membrane protein.
Highly expressed in the cerebellum, particulary in the presynaptic terminals of parallel fibers-Purkinje neurons. Isoform XE and isoform XB are the most abundant isoforms and are detected at low levels in brain and fetal skeletal muscle. The other isoforms are only found at lower levels and not in fetal tissues.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIB subfamily.
Catalyzes the hydrolysis of ATP coupled with the transport of calcium from the cytoplasm to the extracellular space thereby maintaining intracellular calcium homeostasis. Plays a role in blood pressure regulation through regulation of intracellular calcium concentration and nitric oxide production leading to regulation of vascular smooth muscle cells vasoconstriction. Positively regulates bone mineralization through absorption of calcium from the intestine. Plays dual roles in osteoclast differentiation and survival by regulating RANKL-induced calcium oscillations in preosteoclasts and mediating calcium extrusion in mature osteoclasts (By similarity). Regulates insulin sensitivity through calcium/calmodulin signaling pathway by regulating AKT1 activation and NOS3 activation in endothelial cells.
Cell membrane>Multi-pass membrane protein. Basolateral cell membrane. Cell junction>Synapse.
Note: Colocalizes with SV2A in photoreceptor synaptic terminals. Colocalizes with NPTN to the immunological synapse. Colocalizes with EPB41 to the basolateral membrane in enterocyte.
Isoform B: Ubiquitously expressed. Isoform C: Found in brain cortex, skeletal muscle and heart muscle. Isoform D: Has only been found in fetal skeletal muscle. Isoform K: Found in small intestine and liver. Abundantly expressed in the endometrial epithelial cells and glandular epithelial cells in early-proliferative phase and early-secretory phases.
Isoforms A, C, D and E contain an additional calmodulin-binding subdomain B which is different in the different splice variants and shows pH dependent calmodulin binding properties.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIB subfamily.
This magnesium-dependent enzyme catalyzes the hydrolysis of ATP coupled with the transport of calcium out of the cell.
Cell junction>Synapse. Cell membrane>Multi-pass membrane protein.
Mainly expressed in brain cortex. Found in low levels in skeletal muscle, heart muscle, stomach, liver, kidney and lung. Isoforms containing segment B are found in brain cortex and at low levels in other tissues. Isoforms containing segments X and W are found at low levels in all tissues. Isoforms containing segment A and segment Z are found at low levels in skeletal muscle and heart muscle.
Belongs to the cation transport ATPase (P-type) (TC 3.A.3) family. Type IIB subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > Calcium signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cGMP-PKG signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Organismal Systems > Circulatory system > Adrenergic signaling in cardiomyocytes. (View pathway)
· Organismal Systems > Endocrine system > Aldosterone synthesis and secretion.
· Organismal Systems > Excretory system > Endocrine and other factor-regulated calcium reabsorption.
· Organismal Systems > Digestive system > Salivary secretion.
· Organismal Systems > Digestive system > Pancreatic secretion.
· Organismal Systems > Digestive system > Mineral absorption.
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.