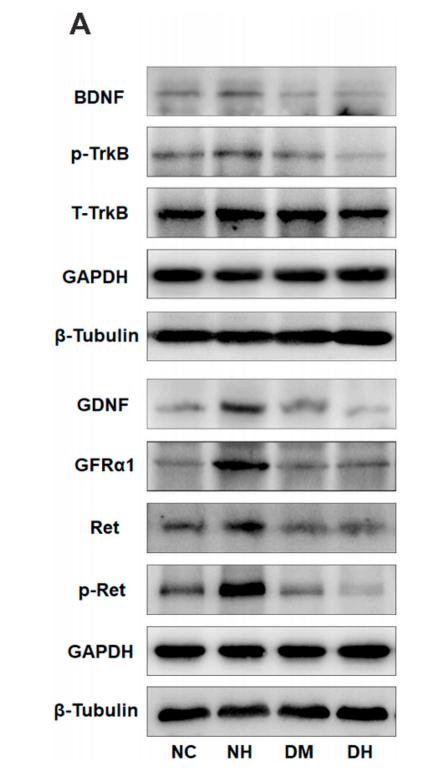
Phospho-Ret (Tyr1062) Antibody - #AF3120
| Product: | Phospho-Ret (Tyr1062) Antibody |
| Catalog: | AF3120 |
| Description: | Rabbit polyclonal antibody to Phospho-Ret (Tyr1062) |
| Application: | WB IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 175kDa; 124kD(Calculated). |
| Uniprot: | P07949 |
| RRID: | AB_2834555 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3120, RRID:AB_2834555.
Fold/Unfold
C ret; Cadherin family member 12; Cadherin related family member 16; CDHF 12; CDHF12; CDHR16; ELKS Fusion gene; HSCR 1; HSCR1; Hydroxyaryl protein kinase; MEN2A; MEN2B; MTC 1; MTC1; Multiple endocrine neoplasia and medullary thyroid carcinoma 1; Oncogene RET; Proto oncogene tyrosine protein kinase receptor ret; Proto-oncogene c-Ret; Proto-oncogene tyrosine-protein kinase receptor ret; PTC; RET; RET ELE1; Ret Proto oncogene; RET transforming sequence; RET_HUMAN; RET51; RET9; tyrosine-protein kinase receptor ret;
Immunogens
A synthesized peptide derived from human Ret around the phosphorylation site of Tyr1062.
- P07949 RET_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAKATSGAAGLRLLLLLLLPLLGKVALGLYFSRDAYWEKLYVDQAAGTPLLYVHALRDAPEEVPSFRLGQHLYGTYRTRLHENNWICIQEDTGLLYLNRSLDHSSWEKLSVRNRGFPLLTVYLKVFLSPTSLREGECQWPGCARVYFSFFNTSFPACSSLKPRELCFPETRPSFRIRENRPPGTFHQFRLLPVQFLCPNISVAYRLLEGEGLPFRCAPDSLEVSTRWALDREQREKYELVAVCTVHAGAREEVVMVPFPVTVYDEDDSAPTFPAGVDTASAVVEFKRKEDTVVATLRVFDADVVPASGELVRRYTSTLLPGDTWAQQTFRVEHWPNETSVQANGSFVRATVHDYRLVLNRNLSISENRTMQLAVLVNDSDFQGPGAGVLLLHFNVSVLPVSLHLPSTYSLSVSRRARRFAQIGKVCVENCQAFSGINVQYKLHSSGANCSTLGVVTSAEDTSGILFVNDTKALRRPKCAELHYMVVATDQQTSRQAQAQLLVTVEGSYVAEEAGCPLSCAVSKRRLECEECGGLGSPTGRCEWRQGDGKGITRNFSTCSPSTKTCPDGHCDVVETQDINICPQDCLRGSIVGGHEPGEPRGIKAGYGTCNCFPEEEKCFCEPEDIQDPLCDELCRTVIAAAVLFSFIVSVLLSAFCIHCYHKFAHKPPISSAEMTFRRPAQAFPVSYSSSGARRPSLDSMENQVSVDAFKILEDPKWEFPRKNLVLGKTLGEGEFGKVVKATAFHLKGRAGYTTVAVKMLKENASPSELRDLLSEFNVLKQVNHPHVIKLYGACSQDGPLLLIVEYAKYGSLRGFLRESRKVGPGYLGSGGSRNSSSLDHPDERALTMGDLISFAWQISQGMQYLAEMKLVHRDLAARNILVAEGRKMKISDFGLSRDVYEEDSYVKRSQGRIPVKWMAIESLFDHIYTTQSDVWSFGVLLWEIVTLGGNPYPGIPPERLFNLLKTGHRMERPDNCSEEMYRLMLQCWKQEPDKRPVFADISKDLEKMMVKRRDYLDLAASTPSDSLIYDDGLSEEETPLVDCNNAPLPRALPSTWIENKLYGMSDPNWPGESPVPLTRADGTNTGFPRYPNDSVYANWMLSPSAAKLMDTFDS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Receptor tyrosine-protein kinase involved in numerous cellular mechanisms including cell proliferation, neuronal navigation, cell migration, and cell differentiation upon binding with glial cell derived neurotrophic factor family ligands. Phosphorylates PTK2/FAK1. Regulates both cell death/survival balance and positional information. Required for the molecular mechanisms orchestration during intestine organogenesis; involved in the development of enteric nervous system and renal organogenesis during embryonic life, and promotes the formation of Peyer's patch-like structures, a major component of the gut-associated lymphoid tissue. Modulates cell adhesion via its cleavage by caspase in sympathetic neurons and mediates cell migration in an integrin (e.g. ITGB1 and ITGB3)-dependent manner. Involved in the development of the neural crest. Active in the absence of ligand, triggering apoptosis through a mechanism that requires receptor intracellular caspase cleavage. Acts as a dependence receptor; in the presence of the ligand GDNF in somatotrophs (within pituitary), promotes survival and down regulates growth hormone (GH) production, but triggers apoptosis in absence of GDNF. Regulates nociceptor survival and size. Triggers the differentiation of rapidly adapting (RA) mechanoreceptors. Mediator of several diseases such as neuroendocrine cancers; these diseases are characterized by aberrant integrins-regulated cell migration. Mediates, through interaction with GDF15-receptor GFRAL, GDF15-induced cell-signaling in the brainstem which induces inhibition of food-intake. Activates MAPK- and AKT-signaling pathways. Isoform 1 in complex with GFRAL induces higher activation of MAPK-signaling pathway than isoform 2 in complex with GFRAL.
Autophosphorylated on C-terminal tyrosine residues upon ligand stimulation. Dephosphorylated by PTPRJ on Tyr-905, Tyr-1015 and Tyr-1062.
Proteolytically cleaved by caspase-3. The soluble RET kinase fragment is able to induce cell death. The extracellular cell-membrane anchored RET cadherin fragment accelerates cell adhesion in sympathetic neurons.
Cell membrane>Single-pass type I membrane protein. Endosome membrane>Single-pass type I membrane protein.
Note: Predominantly located on the plasma membrane. In the presence of SORL1 and GFRA1, directed to endosomes.
Belongs to the protein kinase superfamily. Tyr protein kinase family.
Research Fields
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
References
Application: WB Species: mice Sample: cortex tissue
Application: WB Species: human Sample: SACC-83 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.