Phospho-TAK1 (Thr187) Antibody - #AF3019
| Product: | Phospho-TAK1 (Thr187) Antibody |
| Catalog: | AF3019 |
| Description: | Rabbit polyclonal antibody to Phospho-TAK1 (Thr187) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 70kDa; 67kD(Calculated). |
| Uniprot: | O43318 |
| RRID: | AB_2832996 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF3019, RRID:AB_2832996.
Fold/Unfold
M3K7_HUMAN; MAP3K 7; Map3k7; MEKK7; Mitogen activated protein kinase kinase kinase 7; Mitogen-activated protein kinase kinase kinase 7; TAK1; TGF beta activated kinase 1; TGF-beta-activated kinase 1; TGF1a; Transforming growth factor beta activated kinase 1; Transforming growth factor-beta-activated kinase 1;
Immunogens
A synthesized peptide derived from human TAK1 around the phosphorylation site of Thr187.
Isoform 1A is the most abundant in ovary, skeletal muscle, spleen and blood mononuclear cells. Isoform 1B is highly expressed in brain, kidney and small intestine. Isoform 1C is the major form in prostate. Isoform 1D is the less abundant form.
- O43318 M3K7_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSTASAASSSSSSSAGEMIEAPSQVLNFEEIDYKEIEVEEVVGRGAFGVVCKAKWRAKDVAIKQIESESERKAFIVELRQLSRVNHPNIVKLYGACLNPVCLVMEYAEGGSLYNVLHGAEPLPYYTAAHAMSWCLQCSQGVAYLHSMQPKALIHRDLKPPNLLLVAGGTVLKICDFGTACDIQTHMTNNKGSAAWMAPEVFEGSNYSEKCDVFSWGIILWEVITRRKPFDEIGGPAFRIMWAVHNGTRPPLIKNLPKPIESLMTRCWSKDPSQRPSMEEIVKIMTHLMRYFPGADEPLQYPCQYSDEGQSNSATSTGSFMDIASTNTSNKSDTNMEQVPATNDTIKRLESKLLKNQAKQQSESGRLSLGASRGSSVESLPPTSEGKRMSADMSEIEARIAATTAYSKPKRGHRKTASFGNILDVPEIVISGNGQPRRRSIQDLTVTGTEPGQVSSRSSSPSVRMITTSGPTSEKPTRSHPWTPDDSTDTNGSDNSIPMAYLTLDHQLQPLAPCPNSKESMAVFEQHCKMAQEYMKVQTEIALLLQRKQELVAELDQDEKDQQNTSRLVQEHKKLLDENKSLSTYYQQCKKQLEVIRSQQQKRQGTS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine kinase which acts as an essential component of the MAP kinase signal transduction pathway. Plays an important role in the cascades of cellular responses evoked by changes in the environment. Mediates signal transduction of TRAF6, various cytokines including interleukin-1 (IL-1), transforming growth factor-beta (TGFB), TGFB-related factors like BMP2 and BMP4, toll-like receptors (TLR), tumor necrosis factor receptor CD40 and B-cell receptor (BCR). Ceramides are also able to activate MAP3K7/TAK1. Once activated, acts as an upstream activator of the MKK/JNK signal transduction cascade and the p38 MAPK signal transduction cascade through the phosphorylation and activation of several MAP kinase kinases like MAP2K1/MEK1, MAP2K3/MKK3, MAP2K6/MKK6 and MAP2K7/MKK7. These MAP2Ks in turn activate p38 MAPKs, c-jun N-terminal kinases (JNKs) and I-kappa-B kinase complex (IKK). Both p38 MAPK and JNK pathways control the transcription factors activator protein-1 (AP-1), while nuclear factor-kappa B is activated by IKK. MAP3K7 activates also IKBKB and MAPK8/JNK1 in response to TRAF6 signaling and mediates BMP2-induced apoptosis. In osmotic stress signaling, plays a major role in the activation of MAPK8/JNK1, but not that of NF-kappa-B. Promotes TRIM5 capsid-specific restriction activity. Phosphorylates RIPK1 at 'Ser-321' which positively regulates RIPK1 interaction with RIPK3 to promote necroptosis but negatively regulates RIPK1 kinase activity and its interaction with FADD to mediate apoptosis (By similarity).
Association with TAB1/MAP3K7IP1 promotes autophosphorylation at Ser-192 and subsequent activation. Association with TAB2/MAP3K7IP2, itself associated with free unanchored Lys-63 polyubiquitin chain, promotes autophosphorylation and subsequent activation of MAP3K7. Dephosphorylation at Ser-192 by PPM1B/PP2CB and at Thr-187 by PP2A and PPP6C leads to inactivation.
'Lys-48'-linked polyubiquitination at Lys-72 is induced by TNFalpha, and leads to proteasomal degradation. Undergoes 'Lys-48'-linked polyubiquitination catalyzed by ITCH (By similarity). Requires 'Lys-63'-linked polyubiquitination for autophosphorylation and subsequent activation. 'Lys-63'-linked ubiquitination does not lead to proteasomal degradation. Deubiquitinated by CYLD, a protease that selectively cleaves 'Lys-63'-linked ubiquitin chains. Deubiquitinated by Y.enterocolitica YopP.
(Microbial infection) Cleaved and inactivated by the proteases 3C of coxsackievirus A16 and human enterovirus D68, allowing the virus to disrupt TRAF6-triggered NF-kappa-B induction.
Cytoplasm. Cell membrane>Peripheral membrane protein>Cytoplasmic side.
Note: Although the majority of MAP3K7/TAK1 is found in the cytosol, when complexed with TAB1/MAP3K7IP1 and TAB2/MAP3K7IP2, it is also localized at the cell membrane.
Isoform 1A is the most abundant in ovary, skeletal muscle, spleen and blood mononuclear cells. Isoform 1B is highly expressed in brain, kidney and small intestine. Isoform 1C is the major form in prostate. Isoform 1D is the less abundant form.
Belongs to the protein kinase superfamily. STE Ser/Thr protein kinase family. MAP kinase kinase kinase subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Adherens junction. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TNF signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Parasitic > Leishmaniasis.
· Human Diseases > Infectious diseases: Parasitic > Toxoplasmosis.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > NOD-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > RIG-I-like receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > IL-17 signaling pathway. (View pathway)
· Organismal Systems > Immune system > T cell receptor signaling pathway. (View pathway)
References
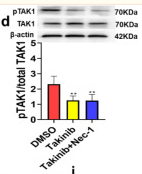
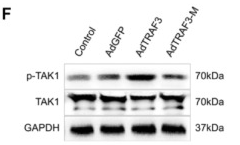
Application: WB Species: Human Sample: neural cells
Application: WB Species: Rat Sample: rMC‐1 cells
Application: WB Species: Rat Sample: NPCs
Application: WB Species: Rat Sample: hippocampal
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.