APBB1IP Antibody - #DF7836
| Product: | APBB1IP Antibody |
| Catalog: | DF7836 |
| Description: | Rabbit polyclonal antibody to APBB1IP |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse |
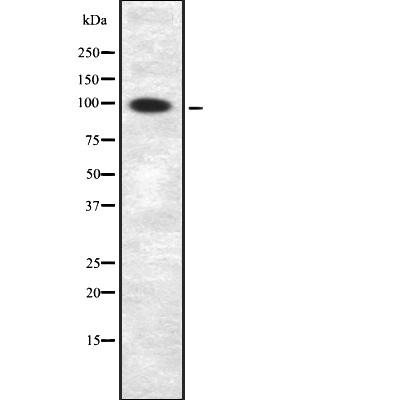
| Mol.Wt.: | 100 kd; 73kD(Calculated). |
| Uniprot: | Q7Z5R6 |
| RRID: | AB_2841276 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7836, RRID:AB_2841276.
Fold/Unfold
AB1IP; Amyloid beta A4 precursor protein binding family B member 1 interacting protein; Amyloid beta precursor protein binding family B member 1; APBB1 interacting protein 1; INAG1; OTTHUMP00000019344; PREL 1; PREL1; Proline rich EVH1 ligand 1; Proline rich protein 73; Rap1 GTP interacting adapter molecule; RAP1 interacting adaptor molecule; RARP 1; RARP1; Retinoic acid responsive proline rich protein 1;
Immunogens
A synthesized peptide derived from human APBB1IP, corresponding to a region within the internal amino acids.
Widely expressed with high expression in thymus, spleen, lymph node, bone marrow and peripheral leukocytes.
- Q7Z5R6 AB1IP_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGESSEDIDQMFSTLLGEMDLLTQSLGVDTLPPPDPNPPRAEFNYSVGFKDLNESLNALEDQDLDALMADLVADISEAEQRTIQAQKESLQNQHHSASLQASIFSGAASLGYGTNVAATGISQYEDDLPPPPADPVLDLPLPPPPPEPLSQEEEEAQAKADKIKLALEKLKEAKVKKLVVKVHMNDNSTKSLMVDERQLARDVLDNLFEKTHCDCNVDWCLYEIYPELQIERFFEDHENVVEVLSDWTRDTENKILFLEKEEKYAVFKNPQNFYLDNRGKKESKETNEKMNAKNKESLLEESFCGTSIIVPELEGALYLKEDGKKSWKRRYFLLRASGIYYVPKGKTKTSRDLACFIQFENVNIYYGTQHKMKYKAPTDYCFVLKHPQIQKESQYIKYLCCDDTRTLNQWVMGIRIAKYGKTLYDNYQRAVAKAGLASRWTNLGTVNAAAPAQPSTGPKTGTTQPNGQIPQATHSVSAVLQEAQRHAETSKDKKPALGNHHDPAVPRAPHAPKSSLPPPPPVRRSSDTSGSPATPLKAKGTGGGGLPAPPDDFLPPPPPPPPLDDPELPPPPPDFMEPPPDFVPPPPPSYAGIAGSELPPPPPPPPAPAPAPVPDSARPPPAVAKRPPVPPKRQENPGHPGGAGGGEQDFMSDLMKALQKKRGNVS
Research Backgrounds
Appears to function in the signal transduction from Ras activation to actin cytoskeletal remodeling. Suppresses insulin-induced promoter activities through AP1 and SRE. Mediates Rap1-induced adhesion.
Cell membrane>Peripheral membrane protein. Cell projection>Lamellipodium. Cell junction>Focal adhesion. Cytoplasm>Cytoskeleton.
Note: Colocalizes with ENA/VASP proteins at lamellipodia tips and focal adhesions, and F-actin at the leading edge. At the membrane surface, associates, via the PH domain, preferentially with the inositol phosphates, PtdIns(5)P and PtdIns(3)P. This binding appears to be necessary for the efficient interaction of the RA domain to Ras-GTPases (By similarity).
Widely expressed with high expression in thymus, spleen, lymph node, bone marrow and peripheral leukocytes.
The two Pro-rich regions are required for the suppression of AP1 transcription activity.
Belongs to the MRL family.
Research Fields
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.