Phospho-ATR (Ser428) Antibody - #DF7512
| Product: | Phospho-ATR (Ser428) Antibody |
| Catalog: | DF7512 |
| Description: | Rabbit polyclonal antibody to Phospho-ATR (Ser428) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Horse, Rabbit, Dog |
| Mol.Wt.: | 300 kDa; 301kD(Calculated). |
| Uniprot: | Q13535 |
| RRID: | AB_2841011 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7512, RRID:AB_2841011.
Fold/Unfold
Ataxia telangiectasia and Rad3 related; Ataxia telangiectasia and Rad3-related protein; ATR; ATR_HUMAN; FCTCS; FRAP Related Protein 1; FRAP-related protein 1; FRP1; MEC1; MEC1 mitosis entry checkpoint 1 homolog; Protein kinase ATR; RAC3; Rad3 related protein; SCKL; SCKL1; Serine/threonine protein kinase ATR; Serine/threonine-protein kinase ATR;
Immunogens
A synthesized peptide derived from human ATR around the phosphorylation site of Ser428.
Ubiquitous, with highest expression in testis. Isoform 2 is found in pancreas, placenta and liver but not in heart, testis and ovary.
- Q13535 ATR_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGEHGLELASMIPALRELGSATPEEYNTVVQKPRQILCQFIDRILTDVNVVAVELVKKTDSQPTSVMLLDFIQHIMKSSPLMFVNVSGSHEAKGSCIEFSNWIITRLLRIAATPSCHLLHKKICEVICSLLFLFKSKSPAIFGVLTKELLQLFEDLVYLHRRNVMGHAVEWPVVMSRFLSQLDEHMGYLQSAPLQLMSMQNLEFIEVTLLMVLTRIIAIVFFRRQELLLWQIGCVLLEYGSPKIKSLAISFLTELFQLGGLPAQPASTFFSSFLELLKHLVEMDTDQLKLYEEPLSKLIKTLFPFEAEAYRNIEPVYLNMLLEKLCVMFEDGVLMRLKSDLLKAALCHLLQYFLKFVPAGYESALQVRKVYVRNICKALLDVLGIEVDAEYLLGPLYAALKMESMEIIEEIQCQTQQENLSSNSDGISPKRRRLSSSLNPSKRAPKQTEEIKHVDMNQKSILWSALKQKAESLQISLEYSGLKNPVIEMLEGIAVVLQLTALCTVHCSHQNMNCRTFKDCQHKSKKKPSVVITWMSLDFYTKVLKSCRSLLESVQKLDLEATIDKVVKIYDALIYMQVNSSFEDHILEDLCGMLSLPWIYSHSDDGCLKLTTFAANLLTLSCRISDSYSPQAQSRCVFLLTLFPRRIFLEWRTAVYNWALQSSHEVIRASCVSGFFILLQQQNSCNRVPKILIDKVKDDSDIVKKEFASILGQLVCTLHGMFYLTSSLTEPFSEHGHVDLFCRNLKATSQHECSSSQLKASVCKPFLFLLKKKIPSPVKLAFIDNLHHLCKHLDFREDETDVKAVLGTLLNLMEDPDKDVRVAFSGNIKHILESLDSEDGFIKELFVLRMKEAYTHAQISRNNELKDTLILTTGDIGRAAKGDLVPFALLHLLHCLLSKSASVSGAAYTEIRALVAAKSVKLQSFFSQYKKPICQFLVESLHSSQMTALPNTPCQNADVRKQDVAHQREMALNTLSEIANVFDFPDLNRFLTRTLQVLLPDLAAKASPAASALIRTLGKQLNVNRREILINNFKYIFSHLVCSCSKDELERALHYLKNETEIELGSLLRQDFQGLHNELLLRIGEHYQQVFNGLSILASFASSDDPYQGPRDIISPELMADYLQPKLLGILAFFNMQLLSSSVGIEDKKMALNSLMSLMKLMGPKHVSSVRVKMMTTLRTGLRFKDDFPELCCRAWDCFVRCLDHACLGSLLSHVIVALLPLIHIQPKETAAIFHYLIIENRDAVQDFLHEIYFLPDHPELKKIKAVLQEYRKETSESTDLQTTLQLSMKAIQHENVDVRIHALTSLKETLYKNQEKLIKYATDSETVEPIISQLVTVLLKGCQDANSQARLLCGECLGELGAIDPGRLDFSTTETQGKDFTFVTGVEDSSFAYGLLMELTRAYLAYADNSRAQDSAAYAIQELLSIYDCREMETNGPGHQLWRRFPEHVREILEPHLNTRYKSSQKSTDWSGVKKPIYLSKLGSNFAEWSASWAGYLITKVRHDLASKIFTCCSIMMKHDFKVTIYLLPHILVYVLLGCNQEDQQEVYAEIMAVLKHDDQHTINTQDIASDLCQLSTQTVFSMLDHLTQWARHKFQALKAEKCPHSKSNRNKVDSMVSTVDYEDYQSVTRFLDLIPQDTLAVASFRSKAYTRAVMHFESFITEKKQNIQEHLGFLQKLYAAMHEPDGVAGVSAIRKAEPSLKEQILEHESLGLLRDATACYDRAIQLEPDQIIHYHGVVKSMLGLGQLSTVITQVNGVHANRSEWTDELNTYRVEAAWKLSQWDLVENYLAADGKSTTWSVRLGQLLLSAKKRDITAFYDSLKLVRAEQIVPLSAASFERGSYQRGYEYIVRLHMLCELEHSIKPLFQHSPGDSSQEDSLNWVARLEMTQNSYRAKEPILALRRALLSLNKRPDYNEMVGECWLQSARVARKAGHHQTAYNALLNAGESRLAELYVERAKWLWSKGDVHQALIVLQKGVELCFPENETPPEGKNMLIHGRAMLLVGRFMEETANFESNAIMKKYKDVTACLPEWEDGHFYLAKYYDKLMPMVTDNKMEKQGDLIRYIVLHFGRSLQYGNQFIYQSMPRMLTLWLDYGTKAYEWEKAGRSDRVQMRNDLGKINKVITEHTNYLAPYQFLTAFSQLISRICHSHDEVFVVLMEIIAKVFLAYPQQAMWMMTAVSKSSYPMRVNRCKEILNKAIHMKKSLEKFVGDATRLTDKLLELCNKPVDGSSSTLSMSTHFKMLKKLVEEATFSEILIPLQSVMIPTLPSILGTHANHASHEPFPGHWAYIAGFDDMVEILASLQKPKKISLKGSDGKFYIMMCKPKDDLRKDCRLMEFNSLINKCLRKDAESRRRELHIRTYAVIPLNDECGIIEWVNNTAGLRPILTKLYKEKGVYMTGKELRQCMLPKSAALSEKLKVFREFLLPRHPPIFHEWFLRTFPDPTSWYSSRSAYCRSTAVMSMVGYILGLGDRHGENILFDSLTGECVHVDFNCLFNKGETFEVPEIVPFRLTHNMVNGMGPMGTEGLFRRACEVTMRLMRDQREPLMSVLKTFLHDPLVEWSKPVKGHSKAPLNETGEVVNEKAKTHVLDIEQRLQGVIKTRNRVTGLPLSIEGHVHYLIQEATDENLLCQMYLGWTPYM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Serine/threonine protein kinase which activates checkpoint signaling upon genotoxic stresses such as ionizing radiation (IR), ultraviolet light (UV), or DNA replication stalling, thereby acting as a DNA damage sensor. Recognizes the substrate consensus sequence [ST]-Q. Phosphorylates BRCA1, CHEK1, MCM2, RAD17, RPA2, SMC1 and p53/TP53, which collectively inhibit DNA replication and mitosis and promote DNA repair, recombination and apoptosis. Phosphorylates 'Ser-139' of histone variant H2AX at sites of DNA damage, thereby regulating DNA damage response mechanism. Required for FANCD2 ubiquitination. Critical for maintenance of fragile site stability and efficient regulation of centrosome duplication.
Phosphorylated; autophosphorylates in vitro.
Nucleus. Nucleus>PML body. Chromosome.
Note: Depending on the cell type, it can also be found in PML nuclear bodies. Recruited to chromatin during S-phase. Redistributes to discrete nuclear foci upon DNA damage, hypoxia or replication fork stalling.
Ubiquitous, with highest expression in testis. Isoform 2 is found in pancreas, placenta and liver but not in heart, testis and ovary.
Belongs to the PI3/PI4-kinase family. ATM subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > p53 signaling pathway. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Genetic Information Processing > Replication and repair > Fanconi anemia pathway.
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
References
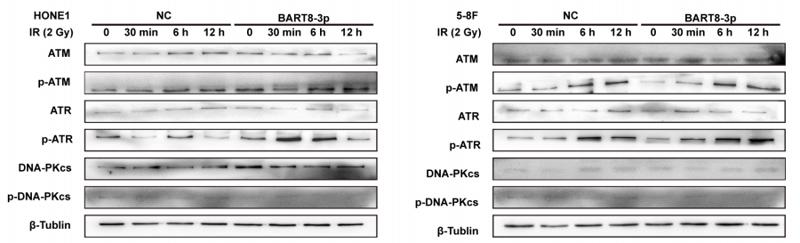
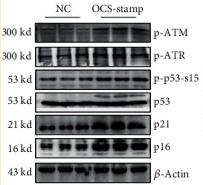
Application: WB Species: mouse Sample: EBV-negative NPC cell
Application: WB Species: Mice Sample: MLE-12 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.