TLR9 Antibody - #DF2970
| Product: | TLR9 Antibody |
| Catalog: | DF2970 |
| Description: | Rabbit polyclonal antibody to TLR9 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Horse, Rabbit, Dog |
| Mol.Wt.: | 117kD,140kD(full), 45kD,75kD(cleaved); 116kD(Calculated). |
| Uniprot: | Q9NR96 |
| RRID: | AB_2840950 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2970, RRID:AB_2840950.
Fold/Unfold
CD 289; CD289; TLR 9; TLR9; TLR9_HUMAN; Toll like receptor 9; Toll like receptor 9 isoform A precursor; Toll like receptor 9 isoform B; Toll-like receptor 9;
Immunogens
A synthesized peptide derived from human TLR9, corresponding to a region within the internal amino acids.
Highly expressed in spleen, lymph node, tonsil and peripheral blood leukocytes, especially in plasmacytoid pre-dendritic cells. Levels are much lower in monocytes and CD11c+ immature dendritic cells. Also detected in lung and liver.
- Q9NR96 TLR9_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MGFCRSALHPLSLLVQAIMLAMTLALGTLPAFLPCELQPHGLVNCNWLFLKSVPHFSMAAPRGNVTSLSLSSNRIHHLHDSDFAHLPSLRHLNLKWNCPPVGLSPMHFPCHMTIEPSTFLAVPTLEELNLSYNNIMTVPALPKSLISLSLSHTNILMLDSASLAGLHALRFLFMDGNCYYKNPCRQALEVAPGALLGLGNLTHLSLKYNNLTVVPRNLPSSLEYLLLSYNRIVKLAPEDLANLTALRVLDVGGNCRRCDHAPNPCMECPRHFPQLHPDTFSHLSRLEGLVLKDSSLSWLNASWFRGLGNLRVLDLSENFLYKCITKTKAFQGLTQLRKLNLSFNYQKRVSFAHLSLAPSFGSLVALKELDMHGIFFRSLDETTLRPLARLPMLQTLRLQMNFINQAQLGIFRAFPGLRYVDLSDNRISGASELTATMGEADGGEKVWLQPGDLAPAPVDTPSSEDFRPNCSTLNFTLDLSRNNLVTVQPEMFAQLSHLQCLRLSHNCISQAVNGSQFLPLTGLQVLDLSHNKLDLYHEHSFTELPRLEALDLSYNSQPFGMQGVGHNFSFVAHLRTLRHLSLAHNNIHSQVSQQLCSTSLRALDFSGNALGHMWAEGDLYLHFFQGLSGLIWLDLSQNRLHTLLPQTLRNLPKSLQVLRLRDNYLAFFKWWSLHFLPKLEVLDLAGNQLKALTNGSLPAGTRLRRLDVSCNSISFVAPGFFSKAKELRELNLSANALKTVDHSWFGPLASALQILDVSANPLHCACGAAFMDFLLEVQAAVPGLPSRVKCGSPGQLQGLSIFAQDLRLCLDEALSWDCFALSLLAVALGLGVPMLHHLCGWDLWYCFHLCLAWLPWRGRQSGRDEDALPYDAFVVFDKTQSAVADWVYNELRGQLEECRGRWALRLCLEERDWLPGKTLFENLWASVYGSRKTLFVLAHTDRVSGLLRASFLLAQQRLLEDRKDVVVLVILSPDGRRSRYVRLRQRLCRQSVLLWPHQPSGQRSFWAQLGMALTRDNHHFYNRNFCQGPTAE
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Key component of innate and adaptive immunity. TLRs (Toll-like receptors) control host immune response against pathogens through recognition of molecular patterns specific to microorganisms. TLR9 is a nucleotide-sensing TLR which is activated by unmethylated cytidine-phosphate-guanosine (CpG) dinucleotides. Acts via MYD88 and TRAF6, leading to NF-kappa-B activation, cytokine secretion and the inflammatory response. Controls lymphocyte response to Helicobacter infection (By similarity). Upon CpG stimulation, induces B-cell proliferation, activation, survival and antibody production.
Activated by proteolytic cleavage of the flexible loop between repeats LRR14 and LRR15 within the ectodomain. Cleavage requires UNC93B1. Proteolytically processed by first removing the majority of the ectodomain by either asparagine endopeptidase (AEP) or a cathepsin followed by a trimming event that is solely cathepsin mediated and required for optimal receptor signaling.
Endoplasmic reticulum membrane>Single-pass type I membrane protein. Endosome. Lysosome. Cytoplasmic vesicle>Phagosome.
Note: Relocalizes from endoplasmic reticulum to endosome and lysosome upon stimulation with agonist. Exit from the ER requires UNC93B1. Endolysosomal localization is required for proteolytic cleavage and subsequent activation. Intracellular localization of the active receptor may prevent from responding to self nucleic acid.
Highly expressed in spleen, lymph node, tonsil and peripheral blood leukocytes, especially in plasmacytoid pre-dendritic cells. Levels are much lower in monocytes and CD11c+ immature dendritic cells. Also detected in lung and liver.
Belongs to the Toll-like receptor family.
Research Fields
· Human Diseases > Infectious diseases: Parasitic > Chagas disease (American trypanosomiasis).
· Human Diseases > Infectious diseases: Parasitic > African trypanosomiasis.
· Human Diseases > Infectious diseases: Parasitic > Malaria.
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Measles.
· Human Diseases > Infectious diseases: Viral > Herpes simplex infection.
· Organismal Systems > Immune system > Toll-like receptor signaling pathway. (View pathway)
References
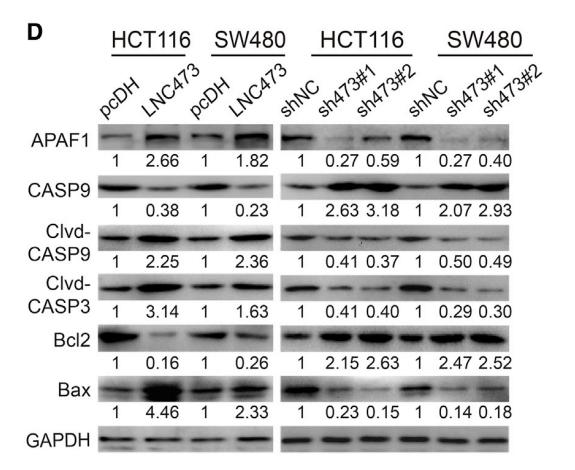
Application: WB Species: human Sample: HUVECs
Application: WB Species: Mice Sample: BMDCs
Application: WB Species: mouse Sample: macrophages
Application: IF/ICC Species: Human Sample: OVISE and CAOV4 cells
Application: WB Species: Rat Sample:
Application: WB Species: Human Sample: HepG2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.