LRP1/CD91 Antibody - #DF2935
| Product: | LRP1/CD91 Antibody |
| Catalog: | DF2935 |
| Description: | Rabbit polyclonal antibody to LRP1/CD91 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Dog |
| Mol.Wt.: | 85kDa; 505kD(Calculated). |
| Uniprot: | Q07954 |
| RRID: | AB_2840921 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2935, RRID:AB_2840921.
Fold/Unfold
A2MR; Alpha 2 macroglobulin receptor; alpha 2MR; Alpha-2-macroglobulin receptor; APOER; Apolipoprotein E receptor; APR; CD 91; CD91; CD91 antigen; IGFBP3R; LDL receptor related protein 1; Low density lipoprotein receptor related protein 1; Low density lipoprotein related protein 1; Low-density lipoprotein receptor-related protein 1 intracellular domain; LRP 1; LRP 515; LRP 85; LRP; LRP ICD; LRP-1; LRP-515; LRP-85; Lrp1; LRP1 protein; LRP1_HUMAN; LRP1A; LRP515; LRP85; LRPICD; MGC88725; Prolow density lipoprotein receptor related protein 1; TbetaR V/LRP 1/IGFBP 3 receptor; TbetaRV/LRP1/IGFBP3 receptor; TGFBR 5; TGFBR5; Type V tgf beta receptor;
Immunogens
A synthesized peptide derived from human LRP1/CD91, corresponding to a region within C-terminal amino acids.
- Q07954 LRP1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MLTPPLLLLLPLLSALVAAAIDAPKTCSPKQFACRDQITCISKGWRCDGERDCPDGSDEAPEICPQSKAQRCQPNEHNCLGTELCVPMSRLCNGVQDCMDGSDEGPHCRELQGNCSRLGCQHHCVPTLDGPTCYCNSSFQLQADGKTCKDFDECSVYGTCSQLCTNTDGSFICGCVEGYLLQPDNRSCKAKNEPVDRPPVLLIANSQNILATYLSGAQVSTITPTSTRQTTAMDFSYANETVCWVHVGDSAAQTQLKCARMPGLKGFVDEHTINISLSLHHVEQMAIDWLTGNFYFVDDIDDRIFVCNRNGDTCVTLLDLELYNPKGIALDPAMGKVFFTDYGQIPKVERCDMDGQNRTKLVDSKIVFPHGITLDLVSRLVYWADAYLDYIEVVDYEGKGRQTIIQGILIEHLYGLTVFENYLYATNSDNANAQQKTSVIRVNRFNSTEYQVVTRVDKGGALHIYHQRRQPRVRSHACENDQYGKPGGCSDICLLANSHKARTCRCRSGFSLGSDGKSCKKPEHELFLVYGKGRPGIIRGMDMGAKVPDEHMIPIENLMNPRALDFHAETGFIYFADTTSYLIGRQKIDGTERETILKDGIHNVEGVAVDWMGDNLYWTDDGPKKTISVARLEKAAQTRKTLIEGKMTHPRAIVVDPLNGWMYWTDWEEDPKDSRRGRLERAWMDGSHRDIFVTSKTVLWPNGLSLDIPAGRLYWVDAFYDRIETILLNGTDRKIVYEGPELNHAFGLCHHGNYLFWTEYRSGSVYRLERGVGGAPPTVTLLRSERPPIFEIRMYDAQQQQVGTNKCRVNNGGCSSLCLATPGSRQCACAEDQVLDADGVTCLANPSYVPPPQCQPGEFACANSRCIQERWKCDGDNDCLDNSDEAPALCHQHTCPSDRFKCENNRCIPNRWLCDGDNDCGNSEDESNATCSARTCPPNQFSCASGRCIPISWTCDLDDDCGDRSDESASCAYPTCFPLTQFTCNNGRCININWRCDNDNDCGDNSDEAGCSHSCSSTQFKCNSGRCIPEHWTCDGDNDCGDYSDETHANCTNQATRPPGGCHTDEFQCRLDGLCIPLRWRCDGDTDCMDSSDEKSCEGVTHVCDPSVKFGCKDSARCISKAWVCDGDNDCEDNSDEENCESLACRPPSHPCANNTSVCLPPDKLCDGNDDCGDGSDEGELCDQCSLNNGGCSHNCSVAPGEGIVCSCPLGMELGPDNHTCQIQSYCAKHLKCSQKCDQNKFSVKCSCYEGWVLEPDGESCRSLDPFKPFIIFSNRHEIRRIDLHKGDYSVLVPGLRNTIALDFHLSQSALYWTDVVEDKIYRGKLLDNGALTSFEVVIQYGLATPEGLAVDWIAGNIYWVESNLDQIEVAKLDGTLRTTLLAGDIEHPRAIALDPRDGILFWTDWDASLPRIEAASMSGAGRRTVHRETGSGGWPNGLTVDYLEKRILWIDARSDAIYSARYDGSGHMEVLRGHEFLSHPFAVTLYGGEVYWTDWRTNTLAKANKWTGHNVTVVQRTNTQPFDLQVYHPSRQPMAPNPCEANGGQGPCSHLCLINYNRTVSCACPHLMKLHKDNTTCYEFKKFLLYARQMEIRGVDLDAPYYNYIISFTVPDIDNVTVLDYDAREQRVYWSDVRTQAIKRAFINGTGVETVVSADLPNAHGLAVDWVSRNLFWTSYDTNKKQINVARLDGSFKNAVVQGLEQPHGLVVHPLRGKLYWTDGDNISMANMDGSNRTLLFSGQKGPVGLAIDFPESKLYWISSGNHTINRCNLDGSGLEVIDAMRSQLGKATALAIMGDKLWWADQVSEKMGTCSKADGSGSVVLRNSTTLVMHMKVYDESIQLDHKGTNPCSVNNGDCSQLCLPTSETTRSCMCTAGYSLRSGQQACEGVGSFLLYSVHEGIRGIPLDPNDKSDALVPVSGTSLAVGIDFHAENDTIYWVDMGLSTISRAKRDQTWREDVVTNGIGRVEGIAVDWIAGNIYWTDQGFDVIEVARLNGSFRYVVISQGLDKPRAITVHPEKGYLFWTEWGQYPRIERSRLDGTERVVLVNVSISWPNGISVDYQDGKLYWCDARTDKIERIDLETGENREVVLSSNNMDMFSVSVFEDFIYWSDRTHANGSIKRGSKDNATDSVPLRTGIGVQLKDIKVFNRDRQKGTNVCAVANGGCQQLCLYRGRGQRACACAHGMLAEDGASCREYAGYLLYSERTILKSIHLSDERNLNAPVQPFEDPEHMKNVIALAFDYRAGTSPGTPNRIFFSDIHFGNIQQINDDGSRRITIVENVGSVEGLAYHRGWDTLYWTSYTTSTITRHTVDQTRPGAFERETVITMSGDDHPRAFVLDECQNLMFWTNWNEQHPSIMRAALSGANVLTLIEKDIRTPNGLAIDHRAEKLYFSDATLDKIERCEYDGSHRYVILKSEPVHPFGLAVYGEHIFWTDWVRRAVQRANKHVGSNMKLLRVDIPQQPMGIIAVANDTNSCELSPCRINNGGCQDLCLLTHQGHVNCSCRGGRILQDDLTCRAVNSSCRAQDEFECANGECINFSLTCDGVPHCKDKSDEKPSYCNSRRCKKTFRQCSNGRCVSNMLWCNGADDCGDGSDEIPCNKTACGVGEFRCRDGTCIGNSSRCNQFVDCEDASDEMNCSATDCSSYFRLGVKGVLFQPCERTSLCYAPSWVCDGANDCGDYSDERDCPGVKRPRCPLNYFACPSGRCIPMSWTCDKEDDCEHGEDETHCNKFCSEAQFECQNHRCISKQWLCDGSDDCGDGSDEAAHCEGKTCGPSSFSCPGTHVCVPERWLCDGDKDCADGADESIAAGCLYNSTCDDREFMCQNRQCIPKHFVCDHDRDCADGSDESPECEYPTCGPSEFRCANGRCLSSRQWECDGENDCHDQSDEAPKNPHCTSQEHKCNASSQFLCSSGRCVAEALLCNGQDDCGDSSDERGCHINECLSRKLSGCSQDCEDLKIGFKCRCRPGFRLKDDGRTCADVDECSTTFPCSQRCINTHGSYKCLCVEGYAPRGGDPHSCKAVTDEEPFLIFANRYYLRKLNLDGSNYTLLKQGLNNAVALDFDYREQMIYWTDVTTQGSMIRRMHLNGSNVQVLHRTGLSNPDGLAVDWVGGNLYWCDKGRDTIEVSKLNGAYRTVLVSSGLREPRALVVDVQNGYLYWTDWGDHSLIGRIGMDGSSRSVIVDTKITWPNGLTLDYVTERIYWADAREDYIEFASLDGSNRHVVLSQDIPHIFALTLFEDYVYWTDWETKSINRAHKTTGTNKTLLISTLHRPMDLHVFHALRQPDVPNHPCKVNNGGCSNLCLLSPGGGHKCACPTNFYLGSDGRTCVSNCTASQFVCKNDKCIPFWWKCDTEDDCGDHSDEPPDCPEFKCRPGQFQCSTGICTNPAFICDGDNDCQDNSDEANCDIHVCLPSQFKCTNTNRCIPGIFRCNGQDNCGDGEDERDCPEVTCAPNQFQCSITKRCIPRVWVCDRDNDCVDGSDEPANCTQMTCGVDEFRCKDSGRCIPARWKCDGEDDCGDGSDEPKEECDERTCEPYQFRCKNNRCVPGRWQCDYDNDCGDNSDEESCTPRPCSESEFSCANGRCIAGRWKCDGDHDCADGSDEKDCTPRCDMDQFQCKSGHCIPLRWRCDADADCMDGSDEEACGTGVRTCPLDEFQCNNTLCKPLAWKCDGEDDCGDNSDENPEECARFVCPPNRPFRCKNDRVCLWIGRQCDGTDNCGDGTDEEDCEPPTAHTTHCKDKKEFLCRNQRCLSSSLRCNMFDDCGDGSDEEDCSIDPKLTSCATNASICGDEARCVRTEKAAYCACRSGFHTVPGQPGCQDINECLRFGTCSQLCNNTKGGHLCSCARNFMKTHNTCKAEGSEYQVLYIADDNEIRSLFPGHPHSAYEQAFQGDESVRIDAMDVHVKAGRVYWTNWHTGTISYRSLPPAAPPTTSNRHRRQIDRGVTHLNISGLKMPRGIAIDWVAGNVYWTDSGRDVIEVAQMKGENRKTLISGMIDEPHAIVVDPLRGTMYWSDWGNHPKIETAAMDGTLRETLVQDNIQWPTGLAVDYHNERLYWADAKLSVIGSIRLNGTDPIVAADSKRGLSHPFSIDVFEDYIYGVTYINNRVFKIHKFGHSPLVNLTGGLSHASDVVLYHQHKQPEVTNPCDRKKCEWLCLLSPSGPVCTCPNGKRLDNGTCVPVPSPTPPPDAPRPGTCNLQCFNGGSCFLNARRQPKCRCQPRYTGDKCELDQCWEHCRNGGTCAASPSGMPTCRCPTGFTGPKCTQQVCAGYCANNSTCTVNQGNQPQCRCLPGFLGDRCQYRQCSGYCENFGTCQMAADGSRQCRCTAYFEGSRCEVNKCSRCLEGACVVNKQSGDVTCNCTDGRVAPSCLTCVGHCSNGGSCTMNSKMMPECQCPPHMTGPRCEEHVFSQQQPGHIASILIPLLLLLLLVLVAGVVFWYKRRVQGAKGFQHQRMTNGAMNVEIGNPTYKMYEGGEPDDVGGLLDADFALDPDKPTNFTNPVYATLYMGGHGSRHSLASTDEKRELLGRGPEDEIGDPLA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Endocytic receptor involved in endocytosis and in phagocytosis of apoptotic cells. Required for early embryonic development. Involved in cellular lipid homeostasis. Involved in the plasma clearance of chylomicron remnants and activated LRPAP1 (alpha 2-macroglobulin), as well as the local metabolism of complexes between plasminogen activators and their endogenous inhibitors. May modulate cellular events, such as APP metabolism, kinase-dependent intracellular signaling, neuronal calcium signaling as well as neurotransmission. Acts as an alpha-2-macroglobulin receptor.
(Microbial infection) Functions as a receptor for Pseudomonas aeruginosa exotoxin A.
Cleaved into a 85 kDa membrane-spanning subunit (LRP-85) and a 515 kDa large extracellular domain (LRP-515) that remains non-covalently associated. Gamma-secretase-dependent cleavage of LRP-85 releases the intracellular domain from the membrane.
The N-terminus is blocked.
Phosphorylated on serine and threonine residues.
Phosphorylated on tyrosine residues upon stimulation with PDGF. Tyrosine phosphorylation promotes interaction with SHC1.
Cell membrane>Single-pass type I membrane protein. Membrane>Coated pit.
Cell membrane>Peripheral membrane protein>Extracellular side. Membrane>Coated pit.
Cytoplasm. Nucleus.
Note: After cleavage, the intracellular domain (LRPICD) is detected both in the cytoplasm and in the nucleus.
Postsynaptic Golgi apparatus. Cytoplasm>Cytoskeleton>Microtubule organizing center.
Note: Localizes to the postsynaptic Golgi apparatus region, also named Golgi outpost, which shapes dendrite morphology by functioning as sites of acentrosomal microtubule nucleation.
Most abundant in liver, brain and lung.
Belongs to the LDLR family.
Research Fields
· Human Diseases > Neurodegenerative diseases > Alzheimer's disease.
· Human Diseases > Infectious diseases: Parasitic > Malaria.
· Organismal Systems > Digestive system > Cholesterol metabolism.
References
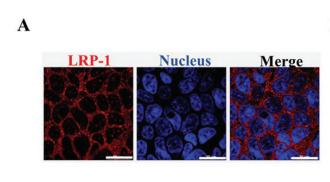
Application: IF/ICC Species: human Sample: Caco-2 cells
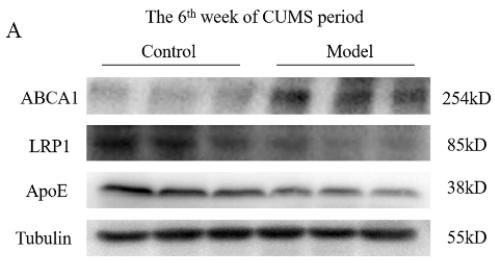
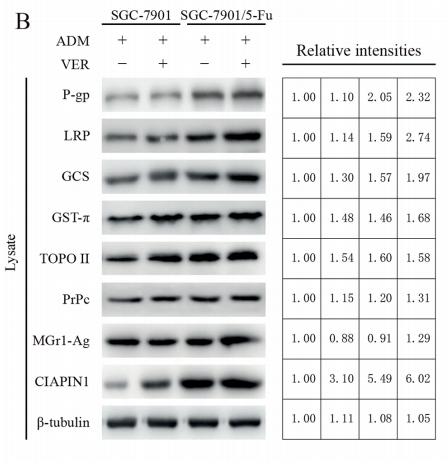
Application: WB Species: Mouse Sample:
Application: WB Species: human Sample: GC cells
Application: WB Species: rat Sample: hippocampus
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.