Bcl6 Antibody - #DF2903
| Product: | Bcl6 Antibody |
| Catalog: | DF2903 |
| Description: | Rabbit polyclonal antibody to Bcl6 |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 78kDa; 79kD(Calculated). |
| Uniprot: | P41182 |
| RRID: | AB_2840892 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF2903, RRID:AB_2840892.
Fold/Unfold
B cell CLL/lymphoma 6; B cell lymphoma 6 protein; B-cell lymphoma 5 protein; B-cell lymphoma 6 protein; BCL 5; Bcl 6; BCL-5; BCL-6; BCL5; BCL6; BCL6_HUMAN; BCL6A; cys his2 zinc finger transcription factor; LAZ 3; LAZ 3 protein; LAZ3; Lymphoma Associated Zinc Finger Gene On Chromosome 3 (LAZ3); Lymphoma associated zinc finger gene on chromosome 3; Protein LAZ-3; ZBTB 27; ZBTB27; Zinc finger and BTB domain containing protein 27; Zinc finger and BTB domain-containing protein 27 (ZBTB27); Zinc finger and BTB domain-containing protein 27; Zinc Finger Protein 51 (ZNF51); Zinc finger protein 51; zinc finger transcription factor BCL6S; ZNF 51; ZNF51;
Immunogens
A synthesized peptide derived from human Bcl6, corresponding to a region within the internal amino acids.
Expressed in germinal center T- and B-cells and in primary immature dendritic cells.
- P41182 BCL6_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MASPADSCIQFTRHASDVLLNLNRLRSRDILTDVVIVVSREQFRAHKTVLMACSGLFYSIFTDQLKCNLSVINLDPEINPEGFCILLDFMYTSRLNLREGNIMAVMATAMYLQMEHVVDTCRKFIKASEAEMVSAIKPPREEFLNSRMLMPQDIMAYRGREVVENNLPLRSAPGCESRAFAPSLYSGLSTPPASYSMYSHLPVSSLLFSDEEFRDVRMPVANPFPKERALPCDSARPVPGEYSRPTLEVSPNVCHSNIYSPKETIPEEARSDMHYSVAEGLKPAAPSARNAPYFPCDKASKEEERPSSEDEIALHFEPPNAPLNRKGLVSPQSPQKSDCQPNSPTESCSSKNACILQASGSPPAKSPTDPKACNWKKYKFIVLNSLNQNAKPEGPEQAELGRLSPRAYTAPPACQPPMEPENLDLQSPTKLSASGEDSTIPQASRLNNIVNRSMTGSPRSSSESHSPLYMHPPKCTSCGSQSPQHAEMCLHTAGPTFPEEMGETQSEYSDSSCENGAFFCNECDCRFSEEASLKRHTLQTHSDKPYKCDRCQASFRYKGNLASHKTVHTGEKPYRCNICGAQFNRPANLKTHTRIHSGEKPYKCETCGARFVQVAHLRAHVLIHTGEKPYPCEICGTRFRHLQTLKSHLRIHTGEKPYHCEKCNLHFRHKSQLRLHLRQKHGAITNTKVQYRVSATDLPPELPKAC
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional repressor mainly required for germinal center (GC) formation and antibody affinity maturation which has different mechanisms of action specific to the lineage and biological functions. Forms complexes with different corepressors and histone deacetylases to repress the transcriptional expression of different subsets of target genes. Represses its target genes by binding directly to the DNA sequence 5'-TTCCTAGAA-3' (BCL6-binding site) or indirectly by repressing the transcriptional activity of transcription factors. In GC B-cells, represses genes that function in differentiation, inflammation, apoptosis and cell cycle control, also autoregulates its transcriptional expression and up-regulates, indirectly, the expression of some genes important for GC reactions, such as AICDA, through the repression of microRNAs expression, like miR155. An important function is to allow GC B-cells to proliferate very rapidly in response to T-cell dependent antigens and tolerate the physiological DNA breaks required for immunglobulin class switch recombination and somatic hypermutation without inducing a p53/TP53-dependent apoptotic response. In follicular helper CD4(+) T-cells (T(FH) cells), promotes the expression of T(FH)-related genes but inhibits the differentiation of T(H)1, T(H)2 and T(H)17 cells. Also required for the establishment and maintenance of immunological memory for both T- and B-cells. Suppresses macrophage proliferation through competition with STAT5 for STAT-binding motifs binding on certain target genes, such as CCL2 and CCND2. In response to genotoxic stress, controls cell cycle arrest in GC B-cells in both p53/TP53-dependedent and -independent manners. Besides, also controls neurogenesis through the alteration of the composition of NOTCH-dependent transcriptional complexes at selective NOTCH targets, such as HES5, including the recruitment of the deacetylase SIRT1 and resulting in an epigenetic silencing leading to neuronal differentiation.
Phosphorylated by MAPK1 in response to antigen receptor activation at Ser-333 and Ser-343. Phosphorylated by ATM in response to genotoxic stress. Phosphorylation induces its degradation by ubiquitin/proteasome pathway.
Polyubiquitinated. Polyubiquitinated by SCF(FBXO11), leading to its degradation by the proteasome. Ubiquitinated by the SCF(FBXL17) complex, leading to its degradation by the proteaseome: ubiquitination by the SCF(FBXL17) complex takes place when aberrant BTB domain dimers are formed.
Acetylated at Lys-379 by EP300 which inhibits the interaction with NuRD complex and the transcriptional repressor function. Deacetylated by HDAC- and SIR2-dependent pathways.
Nucleus.
Expressed in germinal center T- and B-cells and in primary immature dendritic cells.
The BTB domain mediates homodimerization. Its dimer interface mediates peptide binding such as to corepressors BCOR and NCOR2 (PubMed:18212045). Interaction with corepressors through the BTB domain is needed to facilitate the rapid proliferation and survival of GC B-cells but is not involved in the T(FH) formation and BCL6-mediated suppression of T(H)2 and T(H)17 differentiationrequired for GC formation (By similarity).
Research Fields
· Environmental Information Processing > Signal transduction > FoxO signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
References
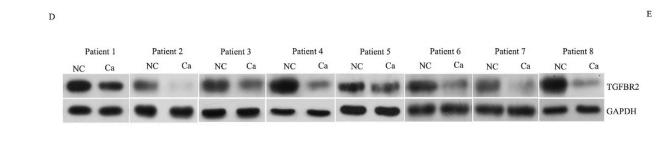
Application: WB Species: human Sample: Gastric cancer cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.