COLEC12 Antibody - #DF10165
| Product: | COLEC12 Antibody |
| Catalog: | DF10165 |
| Description: | Rabbit polyclonal antibody to COLEC12 |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 82 kDa; 82kD(Calculated). |
| Uniprot: | Q5KU26 |
| RRID: | AB_2840744 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF10165, RRID:AB_2840744.
Fold/Unfold
CL P1; CL-P1; CLP1; COL12_HUMAN; COLEC 12; colec12; Collectin placenta 1; Collectin placenta protein 1; Collectin sub family member 12; Collectin-12; hCL P1; hCL-P1; NSR2; Nurse cell scavenger receptor 2; SCARA4; Scavenger receptor class A member 4; Scavenger receptor with C type lectin; Scavenger receptor with C-type lectin; SRCL;
Immunogens
A synthesized peptide derived from human COLEC12, corresponding to a region within C-terminal amino acids.
Expressed in perivascular macrophages. Expressed in plaques-surrounding reactive astrocytes and in perivascular astrocytes associated with cerebral amyloid angiopathy (CAA) in the temporal cortex of Alzheimer patient (at protein level). Strongly expressed in placenta. Moderately expressed in heart, skeletal muscle, small intestine and lung. Weakly expressed in brain, colon, thymus and kidney. Expressed in nurse-like cells. Expressed in reactive astrocytes and vascular/perivascular cells in the brain of Alzheimer patient.
- Q5KU26 COL12_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MKDDFAEEEEVQSFGYKRFGIQEGTQCTKCKNNWALKFSIILLYILCALLTITVAILGYKVVEKMDNVTGGMETSRQTYDDKLTAVESDLKKLGDQTGKKAISTNSELSTFRSDILDLRQQLREITEKTSKNKDTLEKLQASGDALVDRQSQLKETLENNSFLITTVNKTLQAYNGYVTNLQQDTSVLQGNLQNQMYSHNVVIMNLNNLNLTQVQQRNLITNLQRSVDDTSQAIQRIKNDFQNLQQVFLQAKKDTDWLKEKVQSLQTLAANNSALAKANNDTLEDMNSQLNSFTGQMENITTISQANEQNLKDLQDLHKDAENRTAIKFNQLEERFQLFETDIVNIISNISYTAHHLRTLTSNLNEVRTTCTDTLTKHTDDLTSLNNTLANIRLDSVSLRMQQDLMRSRLDTEVANLSVIMEEMKLVDSKHGQLIKNFTILQGPPGPRGPRGDRGSQGPPGPTGNKGQKGEKGEPGPPGPAGERGPIGPAGPPGERGGKGSKGSQGPKGSRGSPGKPGPQGSSGDPGPPGPPGKEGLPGPQGPPGFQGLQGTVGEPGVPGPRGLPGLPGVPGMPGPKGPPGPPGPSGAVVPLALQNEPTPAPEDNGCPPHWKNFTDKCYYFSVEKEIFEDAKLFCEDKSSHLVFINTREEQQWIKKQMVGRESHWIGLTDSERENEWKWLDGTSPDYKNWKAGQPDNWGHGHGPGEDCAGLIYAGQWNDFQCEDVNNFICEKDRETVLSSAL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Scavenger receptor that displays several functions associated with host defense. Promotes binding and phagocytosis of Gram-positive, Gram-negative bacteria and yeast. Mediates the recognition, internalization and degradation of oxidatively modified low density lipoprotein (oxLDL) by vascular endothelial cells. Binds to several carbohydrates including Gal-type ligands, D-galactose, L- and D-fucose, GalNAc, T and Tn antigens in a calcium-dependent manner and internalizes specifically GalNAc in nurse-like cells. Binds also to sialyl Lewis X or a trisaccharide and asialo-orosomucoid (ASOR). May also play a role in the clearance of amyloid-beta in Alzheimer disease.
Membrane>Single-pass type II membrane protein.
Note: Forms clusters on the cell surface.
Expressed in perivascular macrophages. Expressed in plaques-surrounding reactive astrocytes and in perivascular astrocytes associated with cerebral amyloid angiopathy (CAA) in the temporal cortex of Alzheimer patient (at protein level). Strongly expressed in placenta. Moderately expressed in heart, skeletal muscle, small intestine and lung. Weakly expressed in brain, colon, thymus and kidney. Expressed in nurse-like cells. Expressed in reactive astrocytes and vascular/perivascular cells in the brain of Alzheimer patient.
Research Fields
· Cellular Processes > Transport and catabolism > Phagosome. (View pathway)
References
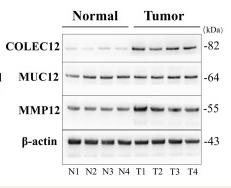
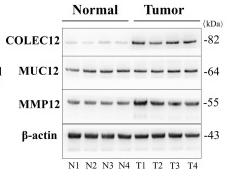
Application: WB Species: Human Sample: liver tissues and HCC tissues
Application: WB Species: Human Sample: HCC tissues and normal liver tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.