Phospho-SYK (Tyr525+Tyr526) Antibody - #AF8404
| Product: | Phospho-SYK (Tyr525+Tyr526) Antibody |
| Catalog: | AF8404 |
| Description: | Rabbit polyclonal antibody to Phospho-SYK (Tyr525+Tyr526) |
| Application: | WB IF/ICC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Chicken |
| Mol.Wt.: | 80kDa; 72kD(Calculated). |
| Uniprot: | P43405 |
| RRID: | AB_2840465 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF8404, RRID:AB_2840465.
Fold/Unfold
EC 2.7.10.2; kinase Syk; KSYK; KSYK_HUMAN; p72-Syk; p72syk; Spleen tyrosine kinase; Syk; Tyrosine protein kinase SYK; Tyrosine-protein kinase SYK;
Immunogens
A synthesized peptide derived from human SYK around the phosphorylation site of Tyr525+Tyr526.
Widely expressed in hematopoietic cells (at protein level) (PubMed:8163536). Expressed in neutrophils (at protein level) (PubMed:15123770). Within the B-cell compartment, expressed from pro- and pre-B cells to plasma cells (PubMed:8163536).
- P43405 KSYK_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MASSGMADSANHLPFFFGNITREEAEDYLVQGGMSDGLYLLRQSRNYLGGFALSVAHGRKAHHYTIERELNGTYAIAGGRTHASPADLCHYHSQESDGLVCLLKKPFNRPQGVQPKTGPFEDLKENLIREYVKQTWNLQGQALEQAIISQKPQLEKLIATTAHEKMPWFHGKISREESEQIVLIGSKTNGKFLIRARDNNGSYALCLLHEGKVLHYRIDKDKTGKLSIPEGKKFDTLWQLVEHYSYKADGLLRVLTVPCQKIGTQGNVNFGGRPQLPGSHPATWSAGGIISRIKSYSFPKPGHRKSSPAQGNRQESTVSFNPYEPELAPWAADKGPQREALPMDTEVYESPYADPEEIRPKEVYLDRKLLTLEDKELGSGNFGTVKKGYYQMKKVVKTVAVKILKNEANDPALKDELLAEANVMQQLDNPYIVRMIGICEAESWMLVMEMAELGPLNKYLQQNRHVKDKNIIELVHQVSMGMKYLEESNFVHRDLAARNVLLVTQHYAKISDFGLSKALRADENYYKAQTHGKWPVKWYAPECINYYKFSSKSDVWSFGVLMWEAFSYGQKPYRGMKGSEVTAMLEKGERMGCPAGCPREMYDLMNLCWTYDVENRPGFAAVELRLRNYYYDVVN
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Non-receptor tyrosine kinase which mediates signal transduction downstream of a variety of transmembrane receptors including classical immunoreceptors like the B-cell receptor (BCR). Regulates several biological processes including innate and adaptive immunity, cell adhesion, osteoclast maturation, platelet activation and vascular development. Assembles into signaling complexes with activated receptors at the plasma membrane via interaction between its SH2 domains and the receptor tyrosine-phosphorylated ITAM domains. The association with the receptor can also be indirect and mediated by adapter proteins containing ITAM or partial hemITAM domains. The phosphorylation of the ITAM domains is generally mediated by SRC subfamily kinases upon engagement of the receptor. More rarely signal transduction via SYK could be ITAM-independent. Direct downstream effectors phosphorylated by SYK include VAV1, PLCG1, PI-3-kinase, LCP2 and BLNK. Initially identified as essential in B-cell receptor (BCR) signaling, it is necessary for the maturation of B-cells most probably at the pro-B to pre-B transition. Activated upon BCR engagement, it phosphorylates and activates BLNK an adapter linking the activated BCR to downstream signaling adapters and effectors. It also phosphorylates and activates PLCG1 and the PKC signaling pathway. It also phosphorylates BTK and regulates its activity in B-cell antigen receptor (BCR)-coupled signaling. In addition to its function downstream of BCR plays also a role in T-cell receptor signaling. Plays also a crucial role in the innate immune response to fungal, bacterial and viral pathogens. It is for instance activated by the membrane lectin CLEC7A. Upon stimulation by fungal proteins, CLEC7A together with SYK activates immune cells inducing the production of ROS. Also activates the inflammasome and NF-kappa-B-mediated transcription of chemokines and cytokines in presence of pathogens. Regulates neutrophil degranulation and phagocytosis through activation of the MAPK signaling cascade (By similarity). Required for the stimulation of neutrophil phagocytosis by IL15. Also mediates the activation of dendritic cells by cell necrosis stimuli. Also involved in mast cells activation. Involved in interleukin-3/IL3-mediated signaling pathway in basophils (By similarity). Also functions downstream of receptors mediating cell adhesion. Relays for instance, integrin-mediated neutrophils and macrophages activation and P-selectin receptor/SELPG-mediated recruitment of leukocytes to inflammatory loci. Plays also a role in non-immune processes. It is for instance involved in vascular development where it may regulate blood and lymphatic vascular separation. It is also required for osteoclast development and function. Functions in the activation of platelets by collagen, mediating PLCG2 phosphorylation and activation. May be coupled to the collagen receptor by the ITAM domain-containing FCER1G. Also activated by the membrane lectin CLEC1B that is required for activation of platelets by PDPN/podoplanin. Involved in platelet adhesion being activated by ITGB3 engaged by fibrinogen. Together with CEACAM20, enhances production of the cytokine CXCL8/IL-8 via the NFKB pathway and may thus have a role in the intestinal immune response (By similarity).
Ubiquitinated by CBLB after BCR activation; which promotes proteasomal degradation.
Autophosphorylated. Phosphorylated on tyrosine residues by LYN following receptors engagement. Phosphorylation on Tyr-323 creates a binding site for CBL, an adapter protein that serves as a negative regulator of BCR-stimulated calcium ion signaling. Phosphorylation at Tyr-348 creates a binding site for VAV1. Phosphorylation on Tyr-348 and Tyr-352 enhances the phosphorylation and activation of phospholipase C-gamma and the early phase of calcium ion mobilization via a phosphoinositide 3-kinase-independent pathway (By similarity). Phosphorylated on tyrosine residues in response to IL15. Phosphorylation on Ser-297 is very common, it peaks 5 minutes after BCR stimulation, and creates a binding site for YWHAG. Phosphorylation at Tyr-630 creates a binding site for BLNK. Dephosphorylated by PTPN6.
Cell membrane. Cytoplasm>Cytosol.
Widely expressed in hematopoietic cells (at protein level). Expressed in neutrophils (at protein level). Within the B-cell compartment, expressed from pro- and pre-B cells to plasma cells.
The SH2 domains mediate the interaction of SYK with the phosphorylated ITAM domains of transmembrane proteins. Some proteins like CLEC1B have a partial ITAM domain (also called hemITAM) containing a single YxxL motif. The interaction with SYK requires CLEC1B homodimerization.
Belongs to the protein kinase superfamily. Tyr protein kinase family. SYK/ZAP-70 subfamily.
Research Fields
· Environmental Information Processing > Signal transduction > NF-kappa B signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Phospholipase D signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Tuberculosis.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Viral carcinogenesis.
· Organismal Systems > Development > Osteoclast differentiation. (View pathway)
· Organismal Systems > Immune system > Platelet activation. (View pathway)
· Organismal Systems > Immune system > Natural killer cell mediated cytotoxicity. (View pathway)
· Organismal Systems > Immune system > B cell receptor signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc epsilon RI signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
References
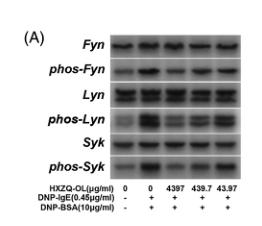
Application: WB Species: Mouse Sample:
Application: WB Species: Mouse Sample: RBL-2H3 cells
Application: WB Species: Mouse Sample: VSMCs
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.