Phospho-Smad1/5/9 (Ser463+Ser465) Antibody - #AF8313
| Product: | Phospho-Smad1/5/9 (Ser463+Ser465) Antibody |
| Catalog: | AF8313 |
| Description: | Rabbit polyclonal antibody to Phospho-Smad1/5/9 (Ser463+Ser465) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Rat, Monkey |
| Prediction: | Pig, Zebrafish, Bovine, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 56kDa; 52kD(Calculated). |
| Uniprot: | Q15797 | Q99717 | O15198 |
| RRID: | AB_2840375 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF8313, RRID:AB_2840375.
Fold/Unfold
BSP-1; BSP1; HsMAD1; JV4-1; JV41; MAD homolog 1; MAD mothers against decapentaplegic homolog 1; Mad related protein 1; Mad-related protein 1; MADH1; MADR1; Mothers against decapentaplegic homolog 1; Mothers against DPP homolog 1; SMA- AND MAD-RELATED PROTEIN 1; SMAD 1; SMAD family member 1; SMAD mothers against DPP homolog 1; Smad1; SMAD1_HUMAN; TGF beta signaling protein 1; Transforming growth factor-beta-signaling protein 1; DKFZp781C1895; DKFZp781O1323; Dwfc; hSmad5; JV5 1; JV5-1; MAD homolog 5; MAD, mothers against decapentaplegic homolog 5; MADH 5; MADH5; Mothers against decapentaplegic homolog 5; mothers against decapentaplegic, drosophila, homolog of, 5; Mothers against DPP homolog 5; MusMLP; SMA and MAD related protein 5; SMAD 5; SMAD family member 5; SMAD, mothers against DPP homolog 5; Smad5; SMAD5_HUMAN; MAD homolog 9; Madh6; Mothers against decapentaplegic; Mothers against decapentaplegic homolog 9; Mothers against DPP homolog 9; SMAD 9; SMAD family member 9; SMAD, mothers against DPP homolog 9 (Drosophila); SMAD8A; SMAD8B; Smad9; SMAD9_HUMAN;
Immunogens
A synthesized peptide derived from human Smad1/5/9 around the phosphorylation site of Ser463+Ser465.
Ubiquitous. Highest expression seen in the heart and skeletal muscle.
Q99717 SMAD5_HUMAN:Ubiquitous.
O15198 SMAD9_HUMAN:Expressed in heart, brain, placenta, lung, skeletal muscle, prostate, testis, ovary and small intestine. Also expressed in fetal brain, lung and kidney.
- Q15797 SMAD1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MNVTSLFSFTSPAVKRLLGWKQGDEEEKWAEKAVDALVKKLKKKKGAMEELEKALSCPGQPSNCVTIPRSLDGRLQVSHRKGLPHVIYCRVWRWPDLQSHHELKPLECCEFPFGSKQKEVCINPYHYKRVESPVLPPVLVPRHSEYNPQHSLLAQFRNLGQNEPHMPLNATFPDSFQQPNSHPFPHSPNSSYPNSPGSSSSTYPHSPTSSDPGSPFQMPADTPPPAYLPPEDPMTQDGSQPMDTNMMAPPLPSEINRGDVQAVAYEEPKHWCSIVYYELNNRVGEAFHASSTSVLVDGFTDPSNNKNRFCLGLLSNVNRNSTIENTRRHIGKGVHLYYVGGEVYAECLSDSSIFVQSRNCNYHHGFHPTTVCKIPSGCSLKIFNNQEFAQLLAQSVNHGFETVYELTKMCTIRMSFVKGWGAEYHRQDVTSTPCWIEIHLHGPLQWLDKVLTQMGSPHNPISSVS
- Q99717 SMAD5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTSMASLFSFTSPAVKRLLGWKQGDEEEKWAEKAVDALVKKLKKKKGAMEELEKALSSPGQPSKCVTIPRSLDGRLQVSHRKGLPHVIYCRVWRWPDLQSHHELKPLDICEFPFGSKQKEVCINPYHYKRVESPVLPPVLVPRHNEFNPQHSLLVQFRNLSHNEPHMPQNATFPDSFHQPNNTPFPLSPNSPYPPSPASSTYPNSPASSGPGSPFQLPADTPPPAYMPPDDQMGQDNSQPMDTSNNMIPQIMPSISSRDVQPVAYEEPKHWCSIVYYELNNRVGEAFHASSTSVLVDGFTDPSNNKSRFCLGLLSNVNRNSTIENTRRHIGKGVHLYYVGGEVYAECLSDSSIFVQSRNCNFHHGFHPTTVCKIPSSCSLKIFNNQEFAQLLAQSVNHGFEAVYELTKMCTIRMSFVKGWGAEYHRQDVTSTPCWIEIHLHGPLQWLDKVLTQMGSPLNPISSVS
- O15198 SMAD9_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MHSTTPISSLFSFTSPAVKRLLGWKQGDEEEKWAEKAVDSLVKKLKKKKGAMDELERALSCPGQPSKCVTIPRSLDGRLQVSHRKGLPHVIYCRVWRWPDLQSHHELKPLECCEFPFGSKQKEVCINPYHYRRVETPVLPPVLVPRHSEYNPQLSLLAKFRSASLHSEPLMPHNATYPDSFQQPPCSALPPSPSHAFSQSPCTASYPHSPGSPSEPESPYQHSVDTPPLPYHATEASETQSGQPVDATADRHVVLSIPNGDFRPVCYEEPQHWCSVAYYELNNRVGETFQASSRSVLIDGFTDPSNNRNRFCLGLLSNVNRNSTIENTRRHIGKGVHLYYVGGEVYAECVSDSSIFVQSRNCNYQHGFHPATVCKIPSGCSLKVFNNQLFAQLLAQSVHHGFEVVYELTKMCTIRMSFVKGWGAEYHRQDVTSTPCWIEIHLHGPLQWLDKVLTQMGSPHNPISSVS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcriptional modulator activated by BMP (bone morphogenetic proteins) type 1 receptor kinase. SMAD1 is a receptor-regulated SMAD (R-SMAD). SMAD1/OAZ1/PSMB4 complex mediates the degradation of the CREBBP/EP300 repressor SNIP1. May act synergistically with SMAD4 and YY1 in bone morphogenetic protein (BMP)-mediated cardiac-specific gene expression.
Phosphorylation of the C-terminal SVS motif by BMP type 1 receptor kinase activates SMAD1 by promoting dissociation from the receptor and trimerization with SMAD4.
Ubiquitinated by SMAD-specific E3 ubiquitin ligase SMURF1, leading to its degradation. Monoubiquitinated, leading to prevent DNA-binding. Deubiquitination by USP15 alleviates inhibition and promotes activation of TGF-beta target genes. Dephosphorylation, probably by PPM1A, induces its export from the nucleus to the cytoplasm (By similarity).
Cytoplasm. Nucleus.
Note: Cytoplasmic in the absence of ligand. Migrates to the nucleus when complexed with SMAD4 (PubMed:15647271). Co-localizes with LEMD3 at the nucleus inner membrane (PubMed:15647271). Exported from the nucleus to the cytoplasm when dephosphorylated (By similarity).
Ubiquitous. Highest expression seen in the heart and skeletal muscle.
The MH2 domain mediates phosphorylation-dependent trimerization through L3 loop binding of phosphoserines in the adjacent subunit.
Belongs to the dwarfin/SMAD family.
Transcriptional modulator activated by BMP (bone morphogenetic proteins) type 1 receptor kinase. SMAD5 is a receptor-regulated SMAD (R-SMAD).
Phosphorylated on serine by BMP (bone morphogenetic proteins) type 1 receptor kinase.
Ubiquitin-mediated proteolysis by SMAD-specific E3 ubiquitin ligase SMURF1.
Cytoplasm. Nucleus.
Note: Cytoplasmic in the absence of ligand. Migrates to the nucleus when complexed with SMAD4.
Ubiquitous.
Belongs to the dwarfin/SMAD family.
Transcriptional modulator activated by BMP (bone morphogenetic proteins) type 1 receptor kinase. SMAD9 is a receptor-regulated SMAD (R-SMAD).
Phosphorylated on serine by BMP (bone morphogenetic proteins) type 1 receptor kinase.
Cytoplasm. Nucleus.
Note: In the cytoplasm in the absence of ligand. Migration to the nucleus when complexed with SMAD4 (By similarity).
Expressed in heart, brain, placenta, lung, skeletal muscle, prostate, testis, ovary and small intestine. Also expressed in fetal brain, lung and kidney.
Belongs to the dwarfin/SMAD family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
References
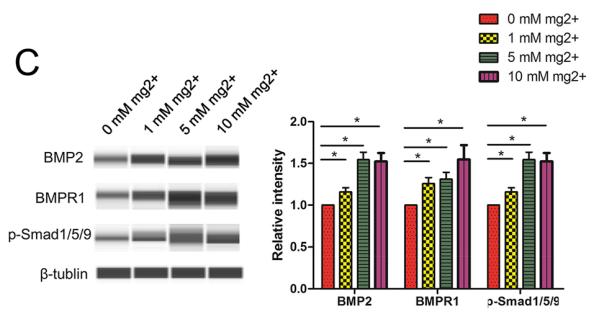
Application: WB Species: Human Sample: DPSCs
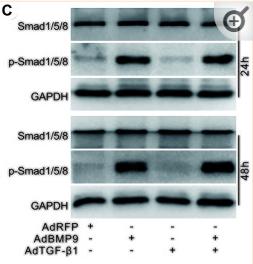
Application: WB Species: Mouse Sample: 3T3-L1 cells
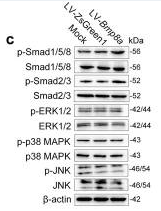
Application: WB Species: Mouse Sample:
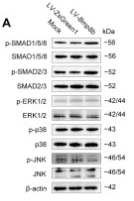
Application: WB Species: Mouse Sample: C3H10T1/2 cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.