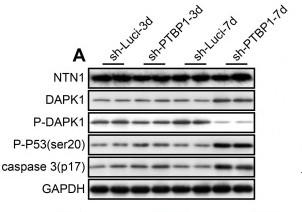
Phospho-DAPK1 (Ser308) Antibody - #AF8046
| Product: | Phospho-DAPK1 (Ser308) Antibody |
| Catalog: | AF8046 |
| Description: | Rabbit polyclonal antibody to Phospho-DAPK1 (Ser308) |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 160kDa; 160kD(Calculated). |
| Uniprot: | P53355 |
| RRID: | AB_2840109 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF8046, RRID:AB_2840109.
Fold/Unfold
DAK1; DAP K1; DAP kinase 1; DAPK 1; DAPK; DAPK1; DAPK1_HUMAN; Death Associated Protein Kinase 1; Death-associated protein kinase 1; DKFZp781I035;
Immunogens
A synthesized peptide derived from human DAPK1 around the phosphorylation site of Ser308.
Isoform 2 is expressed in normal intestinal tissue as well as in colorectal carcinomas.
- P53355 DAPK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MTVFRQENVDDYYDTGEELGSGQFAVVKKCREKSTGLQYAAKFIKKRRTKSSRRGVSREDIEREVSILKEIQHPNVITLHEVYENKTDVILILELVAGGELFDFLAEKESLTEEEATEFLKQILNGVYYLHSLQIAHFDLKPENIMLLDRNVPKPRIKIIDFGLAHKIDFGNEFKNIFGTPEFVAPEIVNYEPLGLEADMWSIGVITYILLSGASPFLGDTKQETLANVSAVNYEFEDEYFSNTSALAKDFIRRLLVKDPKKRMTIQDSLQHPWIKPKDTQQALSRKASAVNMEKFKKFAARKKWKQSVRLISLCQRLSRSFLSRSNMSVARSDDTLDEEDSFVMKAIIHAINDDNVPGLQHLLGSLSNYDVNQPNKHGTPPLLIAAGCGNIQILQLLIKRGSRIDVQDKGGSNAVYWAARHGHVDTLKFLSENKCPLDVKDKSGEMALHVAARYGHADVAQLLCSFGSNPNIQDKEEETPLHCAAWHGYYSVAKALCEAGCNVNIKNREGETPLLTASARGYHDIVECLAEHGADLNACDKDGHIALHLAVRRCQMEVIKTLLSQGCFVDYQDRHGNTPLHVACKDGNMPIVVALCEANCNLDISNKYGRTPLHLAANNGILDVVRYLCLMGASVEALTTDGKTAEDLARSEQHEHVAGLLARLRKDTHRGLFIQQLRPTQNLQPRIKLKLFGHSGSGKTTLVESLKCGLLRSFFRRRRPRLSSTNSSRFPPSPLASKPTVSVSINNLYPGCENVSVRSRSMMFEPGLTKGMLEVFVAPTHHPHCSADDQSTKAIDIQNAYLNGVGDFSVWEFSGNPVYFCCYDYFAANDPTSIHVVVFSLEEPYEIQLNQVIFWLSFLKSLVPVEEPIAFGGKLKNPLQVVLVATHADIMNVPRPAGGEFGYDKDTSLLKEIRNRFGNDLHISNKLFVLDAGASGSKDMKVLRNHLQEIRSQIVSVCPPMTHLCEKIISTLPSWRKLNGPNQLMSLQQFVYDVQDQLNPLASEEDLRRIAQQLHSTGEINIMQSETVQDVLLLDPRWLCTNVLGKLLSVETPRALHHYRGRYTVEDIQRLVPDSDVEELLQILDAMDICARDLSSGTMVDVPALIKTDNLHRSWADEEDEVMVYGGVRIVPVEHLTPFPCGIFHKVQVNLCRWIHQQSTEGDADIRLWVNGCKLANRGAELLVLLVNHGQGIEVQVRGLETEKIKCCLLLDSVCSTIENVMATTLPGLLTVKHYLSPQQLREHHEPVMIYQPRDFFRAQTLKETSLTNTMGGYKESFSSIMCFGCHDVYSQASLGMDIHASDLNLLTRRKLSRLLDPPDPLGKDWCLLAMNLGLPDLVAKYNTSNGAPKDFLPSPLHALLREWTTYPESTVGTLMSKLRELGRRDAADFLLKASSVFKINLDGNGQEAYASSCNSGTSYNSISSVVSR
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Calcium/calmodulin-dependent serine/threonine kinase involved in multiple cellular signaling pathways that trigger cell survival, apoptosis, and autophagy. Regulates both type I apoptotic and type II autophagic cell deaths signal, depending on the cellular setting. The former is caspase-dependent, while the latter is caspase-independent and is characterized by the accumulation of autophagic vesicles. Phosphorylates PIN1 resulting in inhibition of its catalytic activity, nuclear localization, and cellular function. Phosphorylates TPM1, enhancing stress fiber formation in endothelial cells. Phosphorylates STX1A and significantly decreases its binding to STXBP1. Phosphorylates PRKD1 and regulates JNK signaling by binding and activating PRKD1 under oxidative stress. Phosphorylates BECN1, reducing its interaction with BCL2 and BCL2L1 and promoting the induction of autophagy. Phosphorylates TSC2, disrupting the TSC1-TSC2 complex and stimulating mTORC1 activity in a growth factor-dependent pathway. Phosphorylates RPS6, MYL9 and DAPK3. Acts as a signaling amplifier of NMDA receptors at extrasynaptic sites for mediating brain damage in stroke. Cerebral ischemia recruits DAPK1 into the NMDA receptor complex and it phosphorylates GRINB at Ser-1303 inducing injurious Ca(2+) influx through NMDA receptor channels, resulting in an irreversible neuronal death. Required together with DAPK3 for phosphorylation of RPL13A upon interferon-gamma activation which is causing RPL13A involvement in transcript-selective translation inhibition.
Isoform 2 cannot induce apoptosis but can induce membrane blebbing.
Ubiquitinated by the BCR(KLHL20) E3 ubiquitin ligase complex, leading to its degradation by the proteasome.
Removal of the C-terminal tail of isoform 2 (corresponding to amino acids 296-337 of isoform 2) by proteolytic cleavage stimulates maximally its membrane-blebbing function.
In response to mitogenic stimulation (PMA or EGF), phosphorylated at Ser-289; phosphorylation suppresses DAPK1 pro-apoptotic function. Autophosphorylation at Ser-308 inhibits its catalytic activity. Phosphorylation at Ser-734 by MAPK1 increases its catalytic activity and promotes cytoplasmic retention of MAPK1. Endoplasmic-stress can cause dephosphorylation at Ser-308.
Cytoplasm. Cytoplasm>Cytoskeleton.
Note: Colocalizes with MAP1B in the microtubules and cortical actin fibers.
Cytoplasm. Cytoplasm>Cytoskeleton.
Isoform 2 is expressed in normal intestinal tissue as well as in colorectal carcinomas.
The autoinhibitory domain sterically blocks the substrate peptide-binding site by making both hydrophobic and electrostatic contacts with the kinase core.
Belongs to the protein kinase superfamily. CAMK Ser/Thr protein kinase family. DAP kinase subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
References
Application: WB Species: Human Sample: U251 cells
Application: IF/ICC Species: Rat Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.