Phospho-CFTR (Ser737) Antibody - #AF8042
| Product: | Phospho-CFTR (Ser737) Antibody |
| Catalog: | AF8042 |
| Description: | Rabbit polyclonal antibody to Phospho-CFTR (Ser737) |
| Application: | WB IHC |
| Cited expt.: | WB |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken |
| Mol.Wt.: | 168kDa; 168kD(Calculated). |
| Uniprot: | P13569 |
| RRID: | AB_2840105 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF8042, RRID:AB_2840105.
Fold/Unfold
ABC 35; ABC35; ABCC 7; ABCC7; ATP binding cassette sub family C member 7; ATP Binding Cassette Superfamily C Member 7; ATP binding cassette transporter sub family C member 7; ATP-binding cassette sub-family C member 7; cAMP dependent chloride channel; cAMP-dependent chloride channel; CF; CFTR; CFTR/MRP; CFTR_HUMAN; Channel conductance controlling ATPase; Channel conductance-controlling ATPase; Cystic fibrosis transmembrane conductance regulator (ATP-binding cassette sub family C, member 7); Cystic fibrosis transmembrane conductance regulator; Cystic fibrosis transmembrane conductance regulator ATP binding cassette sub family C member 7; Cystic Fibrosis Transmembrane Regulator; dJ760C5.1; MRP 7; MRP7; TNR CFTR;
Immunogens
A synthesized peptide derived from human CFTR around the phosphorylation site of Ser737.
Expressed in the respiratory airway, including bronchial epithelium, and in the female reproductive tract, including oviduct (at protein level) (PubMed:22207244, PubMed:15716351). Detected in pancreatic intercalated ducts in the exocrine tissue, on epithelial cells in intralobular striated ducts in sublingual salivary glands, on apical membranes of crypt cells throughout the small and large intestine, and on the reabsorptive duct in eccrine sweat glands (PubMed:1284548, PubMed:28130590). Detected on the equatorial segment of the sperm head (at protein level) (PubMed:19923167). Detected in nasal and bronchial superficial epithelium (PubMed:15716351). Expressed by the central cells on the sebaceous glands, dermal adipocytes and, at lower levels, by epithelial cells (PubMed:28130590).
- P13569 CFTR_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQRSPLEKASVVSKLFFSWTRPILRKGYRQRLELSDIYQIPSVDSADNLSEKLEREWDRELASKKNPKLINALRRCFFWRFMFYGIFLYLGEVTKAVQPLLLGRIIASYDPDNKEERSIAIYLGIGLCLLFIVRTLLLHPAIFGLHHIGMQMRIAMFSLIYKKTLKLSSRVLDKISIGQLVSLLSNNLNKFDEGLALAHFVWIAPLQVALLMGLIWELLQASAFCGLGFLIVLALFQAGLGRMMMKYRDQRAGKISERLVITSEMIENIQSVKAYCWEEAMEKMIENLRQTELKLTRKAAYVRYFNSSAFFFSGFFVVFLSVLPYALIKGIILRKIFTTISFCIVLRMAVTRQFPWAVQTWYDSLGAINKIQDFLQKQEYKTLEYNLTTTEVVMENVTAFWEEGFGELFEKAKQNNNNRKTSNGDDSLFFSNFSLLGTPVLKDINFKIERGQLLAVAGSTGAGKTSLLMVIMGELEPSEGKIKHSGRISFCSQFSWIMPGTIKENIIFGVSYDEYRYRSVIKACQLEEDISKFAEKDNIVLGEGGITLSGGQRARISLARAVYKDADLYLLDSPFGYLDVLTEKEIFESCVCKLMANKTRILVTSKMEHLKKADKILILHEGSSYFYGTFSELQNLQPDFSSKLMGCDSFDQFSAERRNSILTETLHRFSLEGDAPVSWTETKKQSFKQTGEFGEKRKNSILNPINSIRKFSIVQKTPLQMNGIEEDSDEPLERRLSLVPDSEQGEAILPRISVISTGPTLQARRRQSVLNLMTHSVNQGQNIHRKTTASTRKVSLAPQANLTELDIYSRRLSQETGLEISEEINEEDLKECFFDDMESIPAVTTWNTYLRYITVHKSLIFVLIWCLVIFLAEVAASLVVLWLLGNTPLQDKGNSTHSRNNSYAVIITSTSSYYVFYIYVGVADTLLAMGFFRGLPLVHTLITVSKILHHKMLHSVLQAPMSTLNTLKAGGILNRFSKDIAILDDLLPLTIFDFIQLLLIVIGAIAVVAVLQPYIFVATVPVIVAFIMLRAYFLQTSQQLKQLESEGRSPIFTHLVTSLKGLWTLRAFGRQPYFETLFHKALNLHTANWFLYLSTLRWFQMRIEMIFVIFFIAVTFISILTTGEGEGRVGIILTLAMNIMSTLQWAVNSSIDVDSLMRSVSRVFKFIDMPTEGKPTKSTKPYKNGQLSKVMIIENSHVKKDDIWPSGGQMTVKDLTAKYTEGGNAILENISFSISPGQRVGLLGRTGSGKSTLLSAFLRLLNTEGEIQIDGVSWDSITLQQWRKAFGVIPQKVFIFSGTFRKNLDPYEQWSDQEIWKVADEVGLRSVIEQFPGKLDFVLVDGGCVLSHGHKQLMCLARSVLSKAKILLLDEPSAHLDPVTYQIIRRTLKQAFADCTVILCEHRIEAMLECQQFLVIEENKVRQYDSIQKLLNERSLFRQAISPSDRVKLFPHRNSSKCKSKPQIAALKEETEEEVQDTRL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Epithelial ion channel that plays an important role in the regulation of epithelial ion and water transport and fluid homeostasis. Mediates the transport of chloride ions across the cell membrane. Channel activity is coupled to ATP hydrolysis. The ion channel is also permeable to HCO(3-); selectivity depends on the extracellular chloride concentration. Exerts its function also by modulating the activity of other ion channels and transporters. Plays an important role in airway fluid homeostasis. Contributes to the regulation of the pH and the ion content of the airway surface fluid layer and thereby plays an important role in defense against pathogens. Modulates the activity of the epithelial sodium channel (ENaC) complex, in part by regulating the cell surface expression of the ENaC complex. Inhibits the activity of the ENaC channel containing subunits SCNN1A, SCNN1B and SCNN1G. Inhibits the activity of the ENaC channel containing subunits SCNN1D, SCNN1B and SCNN1G, but not of the ENaC channel containing subunits SCNN1A, SCNN1B and SCNN1G. May regulate bicarbonate secretion and salvage in epithelial cells by regulating the transporter SLC4A7. Can inhibit the chloride channel activity of ANO1. Plays a role in the chloride and bicarbonate homeostasis during sperm epididymal maturation and capacitation.
N-glycosylated.
Phosphorylated; cAMP treatment promotes phosphorylation and activates the channel. Dephosphorylation decreases the ATPase activity (in vitro). Phosphorylation at PKA sites activates the channel. Phosphorylation at PKC sites enhances the response to phosphorylation by PKA. Phosphorylated by AMPK; this inhibits channel activity.
Ubiquitinated, leading to its degradation in the lysosome. Deubiquitination by USP10 in early endosomes enhances its endocytic recycling to the cell membrane. Ubiquitinated by RNF185 during ER stress.
Apical cell membrane>Multi-pass membrane protein. Early endosome membrane>Multi-pass membrane protein. Cell membrane>Multi-pass membrane protein. Recycling endosome membrane>Multi-pass membrane protein. Endoplasmic reticulum membrane>Multi-pass membrane protein. Nucleus.
Note: The channel is internalized from the cell surface into an endosomal recycling compartment, from where it is recycled to the cell membrane (PubMed:17462998, PubMed:19398555, PubMed:20008117). In the oviduct and bronchus, detected on the apical side of epithelial cells, but not associated with cilia (PubMed:22207244). In Sertoli cells, a processed product is detected in the nucleus (By similarity). ER stress induces GORASP2-mediated unconventional (ER/Golgi-independent) trafficking of core-glycosylated CFTR to cell membrane (PubMed:21884936).
Expressed in the respiratory airway, including bronchial epithelium, and in the female reproductive tract, including oviduct (at protein level). Detected in pancreatic intercalated ducts in the exocrine tissue, on epithelial cells in intralobular striated ducts in sublingual salivary glands, on apical membranes of crypt cells throughout the small and large intestine, and on the reabsorptive duct in eccrine sweat glands. Detected on the equatorial segment of the sperm head (at protein level). Detected in nasal and bronchial superficial epithelium. Expressed by the central cells on the sebaceous glands, dermal adipocytes and, at lower levels, by epithelial cells.
Binds and hydrolyzes ATP via the two cytoplasmic ABC transporter nucleotide-binding domains (PubMed:15284228). The two ATP-binding domains interact with each other, forming a head-to-tail dimer (PubMed:17036051). Normal ATPase activity requires interaction between the two domains (PubMed:15284228). The first ABC transporter nucleotide-binding domain has no ATPase activity by itself (By similarity).
The PDZ-binding motif mediates interactions with GOPC and with the SLC4A7, SLC9A3R1/EBP50 complex.
The R region is intrinsically disordered (PubMed:10792060, PubMed:17660831). It mediates channel activation when it is phosphorylated, but not in the absence of phosphorylation (PubMed:10792060).
Belongs to the ABC transporter superfamily. ABCC family. CFTR transporter (TC 3.A.1.202) subfamily.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Tight junction. (View pathway)
· Environmental Information Processing > Membrane transport > ABC transporters.
· Environmental Information Processing > Signal transduction > cAMP signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Vibrio cholerae infection.
· Organismal Systems > Digestive system > Gastric acid secretion.
· Organismal Systems > Digestive system > Pancreatic secretion.
References
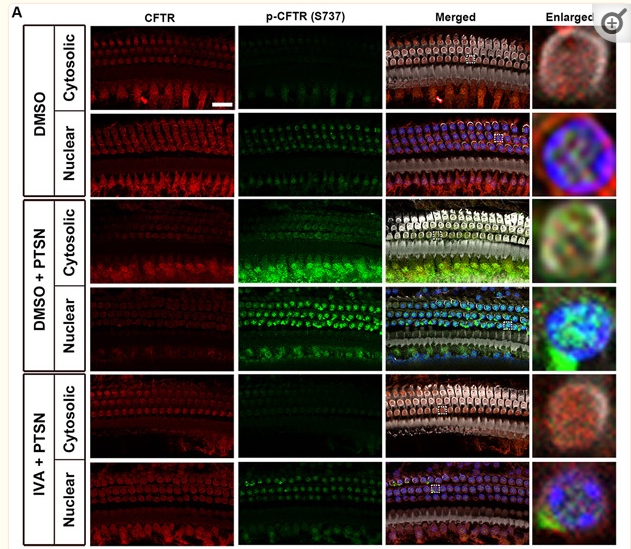
Application: IF/ICC Species: Mouse Sample:
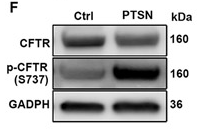
Application: WB Species: Mouse Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.