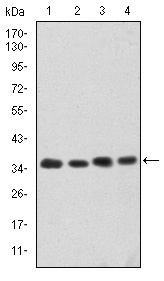
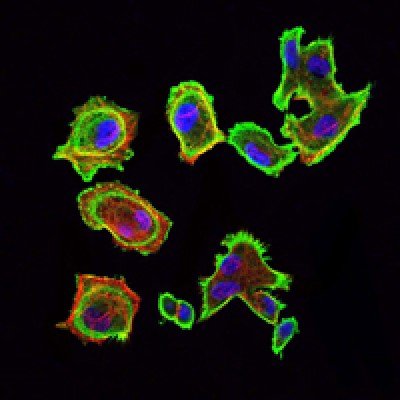
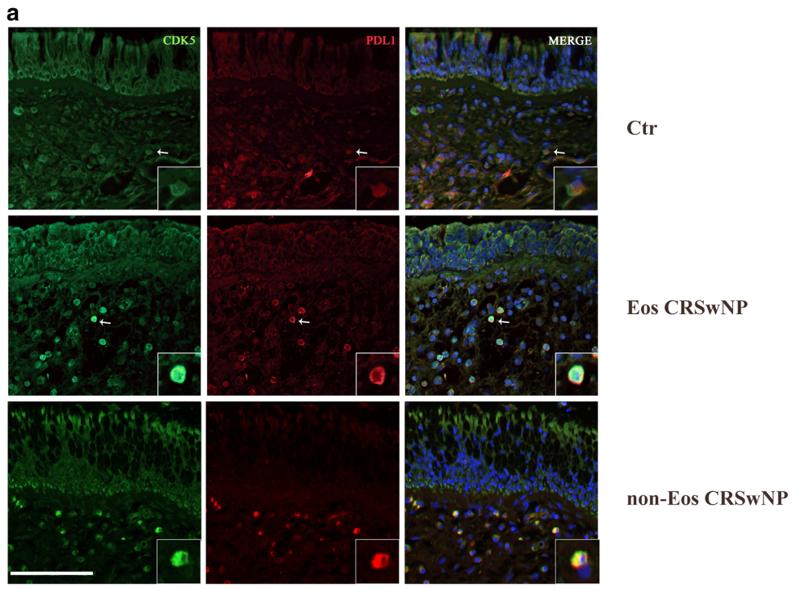
CDK5 Antibody - #BF0121
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# BF0121, RRID:AB_2833913.
Fold/Unfold
Cdk 5; Cdk5; CDK5_HUMAN; Cell division protein kinase 5; Crk6; Cyclin dependent kinase 5; Cyclin-dependent kinase 5; Protein kinase CDK5 splicing; PSSALRE; Serine threonine protein kinase PSSALRE; Serine/threonine-protein kinase PSSALRE; Tau protein kinase II catalytic subunit; TPKII catalytic subunit;
Immunogens
Purified recombinant fragment of human CDK5 expressed in E. Coli.
Isoform 1 is ubiquitously expressed. Accumulates in cortical neurons (at protein level). Isoform 2 has only been detected in testis, skeletal muscle, colon, bone marrow and ovary.
- Q00535 CDK5_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MQKYEKLEKIGEGTYGTVFKAKNRETHEIVALKRVRLDDDDEGVPSSALREICLLKELKHKNIVRLHDVLHSDKKLTLVFEFCDQDLKKYFDSCNGDLDPEIVKSFLFQLLKGLGFCHSRNVLHRDLKPQNLLINRNGELKLADFGLARAFGIPVRCYSAEVVTLWYRPPDVLFGAKLYSTSIDMWSAGCIFAELANAGRPLFPGNDVDDQLKRIFRLLGTPTEEQWPSMTKLPDYKPYPMYPATTSLVNVVPKLNATGRDLLQNLLKCNPVQRISAEEALQHPYFSDFCPP
Research Backgrounds
Proline-directed serine/threonine-protein kinase essential for neuronal cell cycle arrest and differentiation and may be involved in apoptotic cell death in neuronal diseases by triggering abortive cell cycle re-entry. Interacts with D1 and D3-type G1 cyclins. Phosphorylates SRC, NOS3, VIM/vimentin, p35/CDK5R1, MEF2A, SIPA1L1, SH3GLB1, PXN, PAK1, MCAM/MUC18, SEPT5, SYN1, DNM1, AMPH, SYNJ1, CDK16, RAC1, RHOA, CDC42, TONEBP/NFAT5, MAPT/TAU, MAP1B, histone H1, p53/TP53, HDAC1, APEX1, PTK2/FAK1, huntingtin/HTT, ATM, MAP2, NEFH and NEFM. Regulates several neuronal development and physiological processes including neuronal survival, migration and differentiation, axonal and neurite growth, synaptogenesis, oligodendrocyte differentiation, synaptic plasticity and neurotransmission, by phosphorylating key proteins. Activated by interaction with CDK5R1 (p35) and CDK5R2 (p39), especially in post-mitotic neurons, and promotes CDK5R1 (p35) expression in an autostimulation loop. Phosphorylates many downstream substrates such as Rho and Ras family small GTPases (e.g. PAK1, RAC1, RHOA, CDC42) or microtubule-binding proteins (e.g. MAPT/TAU, MAP2, MAP1B), and modulates actin dynamics to regulate neurite growth and/or spine morphogenesis. Phosphorylates also exocytosis associated proteins such as MCAM/MUC18, SEPT5, SYN1, and CDK16/PCTAIRE1 as well as endocytosis associated proteins such as DNM1, AMPH and SYNJ1 at synaptic terminals. In the mature central nervous system (CNS), regulates neurotransmitter movements by phosphorylating substrates associated with neurotransmitter release and synapse plasticity; synaptic vesicle exocytosis, vesicles fusion with the presynaptic membrane, and endocytosis. Promotes cell survival by activating anti-apoptotic proteins BCL2 and STAT3, and negatively regulating of JNK3/MAPK10 activity. Phosphorylation of p53/TP53 in response to genotoxic and oxidative stresses enhances its stabilization by preventing ubiquitin ligase-mediated proteasomal degradation, and induces transactivation of p53/TP53 target genes, thus regulating apoptosis. Phosphorylation of p35/CDK5R1 enhances its stabilization by preventing calpain-mediated proteolysis producing p25/CDK5R1 and avoiding ubiquitin ligase-mediated proteasomal degradation. During aberrant cell-cycle activity and DNA damage, p25/CDK5 activity elicits cell-cycle activity and double-strand DNA breaks that precedes neuronal death by deregulating HDAC1. DNA damage triggered phosphorylation of huntingtin/HTT in nuclei of neurons protects neurons against polyglutamine expansion as well as DNA damage mediated toxicity. Phosphorylation of PXN reduces its interaction with PTK2/FAK1 in matrix-cell focal adhesions (MCFA) during oligodendrocytes (OLs) differentiation. Negative regulator of Wnt/beta-catenin signaling pathway. Activator of the GAIT (IFN-gamma-activated inhibitor of translation) pathway, which suppresses expression of a post-transcriptional regulon of proinflammatory genes in myeloid cells; phosphorylates the linker domain of glutamyl-prolyl tRNA synthetase (EPRS) in a IFN-gamma-dependent manner, the initial event in assembly of the GAIT complex. Phosphorylation of SH3GLB1 is required for autophagy induction in starved neurons. Phosphorylation of TONEBP/NFAT5 in response to osmotic stress mediates its rapid nuclear localization. MEF2 is inactivated by phosphorylation in nucleus in response to neurotoxin, thus leading to neuronal apoptosis. APEX1 AP-endodeoxyribonuclease is repressed by phosphorylation, resulting in accumulation of DNA damage and contributing to neuronal death. NOS3 phosphorylation down regulates NOS3-derived nitrite (NO) levels. SRC phosphorylation mediates its ubiquitin-dependent degradation and thus leads to cytoskeletal reorganization. May regulate endothelial cell migration and angiogenesis via the modulation of lamellipodia formation. Involved in dendritic spine morphogenesis by mediating the EFNA1-EPHA4 signaling. The complex p35/CDK5 participates in the regulation of the circadian clock by modulating the function of CLOCK protein: phosphorylates CLOCK at 'Thr-451' and 'Thr-461' and regulates the transcriptional activity of the CLOCK-ARNTL/BMAL1 heterodimer in association with altered stability and subcellular distribution.
Phosphorylation on Tyr-15 by ABL1 and FYN, and on Ser-159 by casein kinase 1 promotes kinase activity. By contrast, phosphorylation at Thr-14 inhibits activity.
Phosphorylation at Ser-159 is essential for maximal catalytic activity.
Cytoplasm. Cell membrane>Peripheral membrane protein. Perikaryon. Cell projection>Lamellipodium. Cell projection>Growth cone. Cell junction>Synapse>Postsynaptic density.
Note: In axonal growth cone with extension to the peripheral lamellipodia (By similarity). Under neurotoxic stress and neuronal injury conditions, CDK5R (p35) is cleaved by calpain to generate CDK5R1 (p25) in response to increased intracellular calcium. The elevated level of p25, when in complex with CDK5, leads to its subcellular misallocation as well as its hyperactivation. Colocalizes with CTNND2 in the cell body of neuronal cells, and with CTNNB1 in the cell-cell contacts and plasma membrane of undifferentiated and differentiated neuroblastoma cells. Reversibly attached to the plasma membrane in an inactive form when complexed to dephosphorylated p35 or CDK5R2 (p39), p35 phosphorylation releases this attachment and activates CDK5.
Nucleus.
Isoform 1 is ubiquitously expressed. Accumulates in cortical neurons (at protein level). Isoform 2 has only been detected in testis, skeletal muscle, colon, bone marrow and ovary.
Belongs to the protein kinase superfamily. CMGC Ser/Thr protein kinase family. CDC2/CDKX subfamily.
Research Fields
· Human Diseases > Neurodegenerative diseases > Alzheimer's disease.
· Human Diseases > Substance dependence > Cocaine addiction.
· Organismal Systems > Development > Axon guidance. (View pathway)
References
Application: IF/ICC Species: human Sample: nasal mucosal
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.