ALDH1A1 Antibody - #BF0220
| Product: | ALDH1A1 Antibody |
| Catalog: | BF0220 |
| Description: | Mouse monoclonal antibody to ALDH1A1 |
| Application: | WB IHC ELISA |
| Reactivity: | Human |
| Mol.Wt.: | 55kDa; 55kD(Calculated). |
| Uniprot: | P00352 |
| RRID: | AB_2833841 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# BF0220, RRID:AB_2833841.
Fold/Unfold
Acetaldehyde dehydrogenase 1; AHD2; AL1A1_HUMAN; ALDC; Aldehyde dehydrogenase 1 family member A1; Aldehyde dehydrogenase 1 soluble; Aldehyde dehydrogenase 1A1; Aldehyde dehydrogenase; Aldehyde dehydrogenase cytosolic; Aldehyde dehydrogenase family 1 member A1; Aldehyde dehydrogenase liver cytosolic; ALDH 1; ALDH 1A1; ALDH class 1; ALDH, liver cytosolic; ALDH-E1; ALDH1 A1; ALDH1; ALDH11; ALDH1A1; ALHDII; cytosolic; epididymis luminal protein 12; epididymis luminal protein 9; epididymis secretory sperm binding protein Li 53e; HEL-S-53e; MGC2318; PUMB1; RALDH 1; RalDH1; Retinal dehydrogenase 1;
Immunogens
Purified recombinant fragment of human ALDH1A1 expressed in E. Coli.
- P00352 AL1A1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSSSGTPDLPVLLTDLKIQYTKIFINNEWHDSVSGKKFPVFNPATEEELCQVEEGDKEDVDKAVKAARQAFQIGSPWRTMDASERGRLLYKLADLIERDRLLLATMESMNGGKLYSNAYLNDLAGCIKTLRYCAGWADKIQGRTIPIDGNFFTYTRHEPIGVCGQIIPWNFPLVMLIWKIGPALSCGNTVVVKPAEQTPLTALHVASLIKEAGFPPGVVNIVPGYGPTAGAAISSHMDIDKVAFTGSTEVGKLIKEAAGKSNLKRVTLELGGKSPCIVLADADLDNAVEFAHHGVFYHQGQCCIAASRIFVEESIYDEFVRRSVERAKKYILGNPLTPGVTQGPQIDKEQYDKILDLIESGKKEGAKLECGGGPWGNKGYFVQPTVFSNVTDEMRIAKEEIFGPVQQIMKFKSLDDVIKRANNTFYGLSAGVFTKDIDKAITISSALQAGTVWVNCYGVVSAQCPFGGFKMSGNGRELGEYGFHEYTEVKTVTVKISQKNS
PTMs - P00352 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S2 | Acetylation | Uniprot | |
| S2 | Phosphorylation | Uniprot | |
| T6 | Phosphorylation | Uniprot | |
| K22 | Acetylation | Uniprot | |
| K65 | Acetylation | Uniprot | |
| R68 | Methylation | Uniprot | |
| R85 | Methylation | Uniprot | |
| K91 | Acetylation | Uniprot | |
| T105 | Phosphorylation | Uniprot | |
| Y119 | Phosphorylation | Uniprot | |
| K128 | Acetylation | Uniprot | |
| K128 | Ubiquitination | Uniprot | |
| K139 | Acetylation | Uniprot | |
| K139 | Ubiquitination | Uniprot | |
| T144 | Phosphorylation | Uniprot | |
| Y154 | Phosphorylation | Uniprot | |
| K252 | Acetylation | Uniprot | |
| T267 | Phosphorylation | Uniprot | |
| S314 | Phosphorylation | Uniprot | |
| Y316 | Phosphorylation | Uniprot | |
| T337 | Phosphorylation | Uniprot | |
| T341 | Phosphorylation | Uniprot | |
| K353 | Acetylation | Uniprot | |
| S360 | Phosphorylation | Uniprot | |
| K362 | Acetylation | Uniprot | |
| K367 | Acetylation | Uniprot | |
| K398 | Ubiquitination | Uniprot | |
| K410 | Acetylation | Uniprot | |
| K412 | Acetylation | Uniprot | |
| K412 | Ubiquitination | Uniprot | |
| S413 | Phosphorylation | Uniprot | |
| K419 | Acetylation | Uniprot | |
| T424 | Phosphorylation | Uniprot | |
| Y426 | Phosphorylation | Uniprot | |
| S429 | Phosphorylation | Uniprot | |
| K435 | Acetylation | Uniprot | |
| K435 | Ubiquitination | Uniprot | |
| T442 | Phosphorylation | Uniprot | |
| Y481 | Phosphorylation | Uniprot | |
| Y486 | Phosphorylation | Uniprot | |
| K490 | Ubiquitination | Uniprot | |
| T493 | Phosphorylation | Uniprot | |
| K495 | Acetylation | Uniprot | |
| K495 | Ubiquitination | Uniprot |
Research Backgrounds
Can convert/oxidize retinaldehyde to retinoic acid. Binds free retinal and cellular retinol-binding protein-bound retinal (By similarity). May have a broader specificity and oxidize other aldehydes in vivo.
The N-terminus is blocked most probably by acetylation.
Cytoplasm>Cytosol.
Homotetramer.
Belongs to the aldehyde dehydrogenase family.
Research Fields
· Metabolism > Metabolism of cofactors and vitamins > Retinol metabolism.
· Metabolism > Global and overview maps > Metabolic pathways.
References
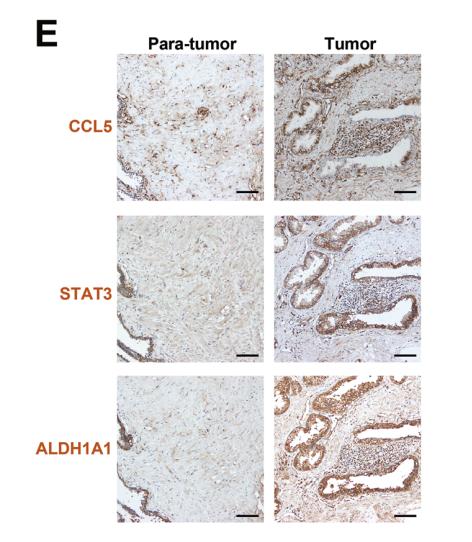
Application: IHC Species: human Sample: prostate cancer cells
Application: IF/ICC Species: mouse Sample: 4T1-Luc xenografts
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.