PGK1 Antibody - #DF6722
| Product: | PGK1 Antibody |
| Catalog: | DF6722 |
| Description: | Rabbit polyclonal antibody to PGK1 |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog |
| Mol.Wt.: | 45kDa; 45kD(Calculated). |
| Uniprot: | P00558 |
| RRID: | AB_2838684 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF6722, RRID:AB_2838684.
Fold/Unfold
Cell migration-inducing gene 10 protein; Epididymis secretory sperm binding protein Li 68p; HEL S 68p; MGC117307; MGC8947; MIG10; pgk1; PGK1_HUMAN; PGKA; Phosphoglycerate kinase 1; Primer recognition protein 2; PRP 2;
Immunogens
Mainly expressed in spermatogonia. Localized on the principle piece in the sperm (at protein level). Expression significantly decreased in the testis of elderly men.
- P00558 PGK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSLSNKLTLDKLDVKGKRVVMRVDFNVPMKNNQITNNQRIKAAVPSIKFCLDNGAKSVVLMSHLGRPDGVPMPDKYSLEPVAVELKSLLGKDVLFLKDCVGPEVEKACANPAAGSVILLENLRFHVEEEGKGKDASGNKVKAEPAKIEAFRASLSKLGDVYVNDAFGTAHRAHSSMVGVNLPQKAGGFLMKKELNYFAKALESPERPFLAILGGAKVADKIQLINNMLDKVNEMIIGGGMAFTFLKVLNNMEIGTSLFDEEGAKIVKDLMSKAEKNGVKITLPVDFVTADKFDENAKTGQATVASGIPAGWMGLDCGPESSKKYAEAVTRAKQIVWNGPVGVFEWEAFARGTKALMDEVVKATSRGCITIIGGGDTATCCAKWNTEDKVSHVSTGGGASLELLEGKVLPGVDALSNI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P00558 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S2 | Acetylation | Uniprot | |
| S2 | Phosphorylation | Uniprot | |
| S4 | Phosphorylation | Uniprot | |
| K6 | Acetylation | Uniprot | |
| K6 | Ubiquitination | Uniprot | |
| K11 | Acetylation | Uniprot | |
| K11 | Ubiquitination | Uniprot | |
| K15 | Ubiquitination | Uniprot | |
| K17 | Ubiquitination | Uniprot | |
| K30 | Acetylation | Uniprot | |
| K30 | Methylation | Uniprot | |
| K30 | Sumoylation | Uniprot | |
| K30 | Ubiquitination | Uniprot | |
| T35 | Phosphorylation | Uniprot | |
| R39 | Methylation | Uniprot | |
| K41 | Ubiquitination | Uniprot | |
| S46 | Phosphorylation | Uniprot | |
| K48 | Acetylation | Uniprot | |
| K48 | Ubiquitination | Uniprot | |
| C50 | S-Nitrosylation | Uniprot | |
| K56 | Acetylation | Uniprot | |
| K56 | Ubiquitination | Uniprot | |
| K75 | Acetylation | Uniprot | |
| K75 | Ubiquitination | Uniprot | |
| Y76 | Phosphorylation | Uniprot | |
| S77 | Phosphorylation | Uniprot | |
| K86 | Acetylation | Uniprot | |
| K86 | Ubiquitination | Uniprot | |
| S87 | Phosphorylation | Uniprot | |
| K91 | Acetylation | Uniprot | |
| K91 | Ubiquitination | Uniprot | |
| K97 | Acetylation | Uniprot | |
| K97 | Ubiquitination | Uniprot | |
| C99 | S-Nitrosylation | Uniprot | |
| K106 | Acetylation | Uniprot | |
| K106 | Ubiquitination | Uniprot | |
| C108 | S-Nitrosylation | Uniprot | |
| S115 | Phosphorylation | Uniprot | |
| R123 | Methylation | Uniprot | |
| K131 | Acetylation | Uniprot | |
| K131 | Ubiquitination | Uniprot | |
| K133 | Acetylation | Uniprot | |
| S136 | Phosphorylation | Uniprot | |
| K139 | Ubiquitination | Uniprot | |
| K141 | Acetylation | Uniprot | |
| K141 | Ubiquitination | Uniprot | |
| K146 | Acetylation | Uniprot | |
| K146 | Ubiquitination | Uniprot | |
| S153 | Phosphorylation | Uniprot | |
| S155 | Phosphorylation | Uniprot | |
| K156 | Acetylation | Uniprot | |
| K156 | Ubiquitination | Uniprot | |
| Y161 | Phosphorylation | Uniprot | |
| T168 | Phosphorylation | Uniprot | |
| S175 | Phosphorylation | Uniprot | |
| K184 | Ubiquitination | Uniprot | |
| K191 | Acetylation | Uniprot | |
| K191 | Methylation | Uniprot | |
| K191 | Ubiquitination | Uniprot | |
| K192 | Ubiquitination | Uniprot | |
| Y196 | Phosphorylation | Uniprot | |
| K199 | Acetylation | Uniprot | |
| K199 | Ubiquitination | Uniprot | |
| S203 | Phosphorylation | Uniprot | |
| K216 | Acetylation | Uniprot | |
| K216 | Ubiquitination | Uniprot | |
| K220 | Acetylation | Uniprot | |
| K220 | Ubiquitination | Uniprot | |
| T243 | Phosphorylation | Uniprot | |
| S256 | Phosphorylation | Uniprot | |
| K264 | Acetylation | Uniprot | |
| K264 | Ubiquitination | Uniprot | |
| K267 | Acetylation | Uniprot | |
| K267 | Ubiquitination | Uniprot | |
| S271 | Phosphorylation | Uniprot | |
| K272 | Acetylation | Uniprot | |
| K272 | Ubiquitination | Uniprot | |
| K275 | Acetylation | Uniprot | |
| K279 | Ubiquitination | Uniprot | |
| T281 | Phosphorylation | Uniprot | |
| T288 | Phosphorylation | Uniprot | |
| K291 | Acetylation | Uniprot | |
| K291 | Ubiquitination | Uniprot | |
| K297 | Ubiquitination | Uniprot | |
| S305 | Phosphorylation | Uniprot | |
| K322 | Acetylation | Uniprot | |
| K322 | Ubiquitination | Uniprot | |
| K323 | Acetylation | Uniprot | |
| K323 | Ubiquitination | Uniprot | |
| K332 | Ubiquitination | Uniprot | |
| K353 | Acetylation | Uniprot | |
| K353 | Ubiquitination | Uniprot | |
| K361 | Acetylation | Uniprot | |
| K361 | Ubiquitination | Uniprot | |
| R365 | Methylation | Uniprot | |
| C367 | S-Nitrosylation | Uniprot | |
| T378 | Phosphorylation | Uniprot | |
| C379 | S-Nitrosylation | Uniprot | |
| C380 | S-Nitrosylation | Uniprot | |
| K382 | Acetylation | Uniprot | |
| K382 | Ubiquitination | Uniprot | |
| K388 | Acetylation | Uniprot | |
| K388 | Ubiquitination | Uniprot | |
| S399 | Phosphorylation | Uniprot | |
| K406 | Acetylation | Uniprot | |
| K406 | Ubiquitination | Uniprot | |
| S415 | Phosphorylation | Uniprot |
Research Backgrounds
Catalyzes one of the two ATP producing reactions in the glycolytic pathway via the reversible conversion of 1,3-diphosphoglycerate to 3-phosphoglycerate. In addition to its role as a glycolytic enzyme, it seems that PGK-1 acts as a polymerase alpha cofactor protein (primer recognition protein). May play a role in sperm motility.
Cytoplasm.
Mainly expressed in spermatogonia. Localized on the principle piece in the sperm (at protein level). Expression significantly decreased in the testis of elderly men.
Monomer.
Belongs to the phosphoglycerate kinase family.
Research Fields
· Environmental Information Processing > Signal transduction > HIF-1 signaling pathway. (View pathway)
· Metabolism > Carbohydrate metabolism > Glycolysis / Gluconeogenesis.
· Metabolism > Global and overview maps > Metabolic pathways.
· Metabolism > Global and overview maps > Carbon metabolism.
· Metabolism > Global and overview maps > Biosynthesis of amino acids.
References
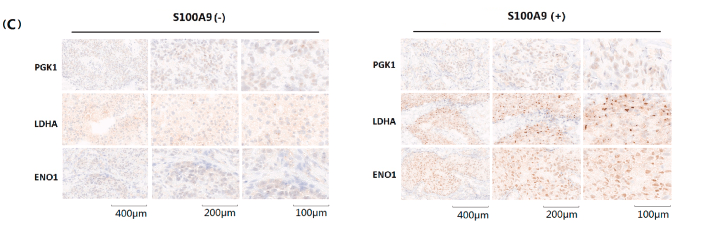
Application: IHC Species: Human Sample: HER2+ BRCA tissues
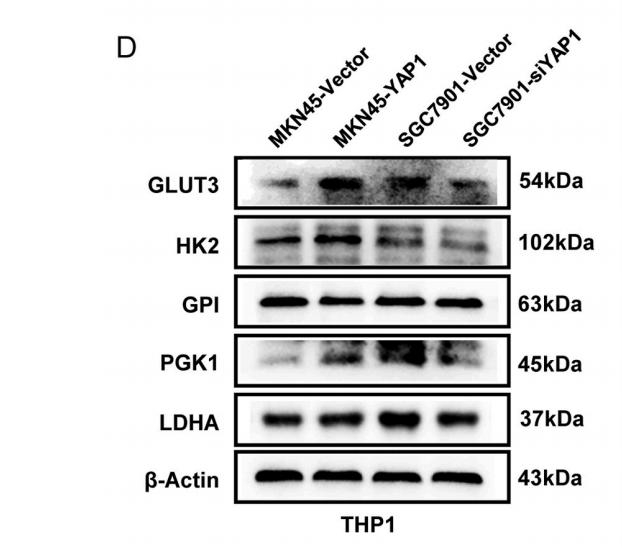
Application: WB Species: Human Sample: Gastric cancer (GC) cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.