Caveolin 1 Antibody - #AF0126
| Product: | Caveolin 1 Antibody |
| Catalog: | AF0126 |
| Description: | Rabbit polyclonal antibody to Caveolin 1 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Horse |
| Mol.Wt.: | 25kDa; 20kD(Calculated). |
| Uniprot: | Q03135 |
| RRID: | AB_2833310 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0126, RRID:AB_2833310.
Fold/Unfold
BSCL3; CAV; CAV1; CAV1_HUMAN; caveolae protein, 22 kD; caveolin 1 alpha isoform; caveolin 1 beta isoform; Caveolin 1 caveolae protein 22kDa; Caveolin-1; Caveolin1; cell growth-inhibiting protein 32; CGL3; LCCNS; MSTP085; OTTHUMP00000025031; PPH3; VIP 21; VIP21;
Immunogens
Skeletal muscle, liver, stomach, lung, kidney and heart (at protein level). Expressed in the brain.
- Q03135 CAV1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MSGGKYVDSEGHLYTVPIREQGNIYKPNNKAMADELSEKQVYDAHTKEIDLVNRDPKHLNDDVVKIDFEDVIAEPEGTHSFDGIWKASFTTFTVTKYWFYRLLSALFGIPMALIWGIYFAILSFLHIWAVVPCIKSFLIEIQCISRVYSIYVHTVCDPLFEAVGKIFSNVRINLQKEI
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q03135 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S2 | Acetylation | Uniprot | |
| S2 | Phosphorylation | Uniprot | |
| K5 | Acetylation | Uniprot | |
| K5 | Ubiquitination | Uniprot | |
| Y6 | Phosphorylation | Uniprot | |
| S9 | Phosphorylation | Uniprot | |
| Y14 | Phosphorylation | P12931 (SRC) , P06241 (FYN) , P00519 (ABL1) | Uniprot |
| T15 | Phosphorylation | Uniprot | |
| Y25 | Phosphorylation | Uniprot | |
| K26 | Sumoylation | Uniprot | |
| K26 | Ubiquitination | Uniprot | |
| K30 | Ubiquitination | Uniprot | |
| S37 | Phosphorylation | Uniprot | |
| K39 | Ubiquitination | Uniprot | |
| Y42 | Phosphorylation | Uniprot | |
| T46 | Phosphorylation | Uniprot | |
| K47 | Ubiquitination | Uniprot | |
| K57 | Acetylation | Uniprot | |
| K57 | Ubiquitination | Uniprot | |
| K65 | Ubiquitination | Uniprot | |
| S80 | Phosphorylation | Uniprot | |
| S88 | Phosphorylation | Uniprot | |
| S168 | Phosphorylation | Uniprot | |
| K176 | Ubiquitination | Uniprot |
Research Backgrounds
May act as a scaffolding protein within caveolar membranes. Forms a stable heterooligomeric complex with CAV2 that targets to lipid rafts and drives caveolae formation. Mediates the recruitment of CAVIN proteins (CAVIN1/2/3/4) to the caveolae. Interacts directly with G-protein alpha subunits and can functionally regulate their activity (By similarity). Involved in the costimulatory signal essential for T-cell receptor (TCR)-mediated T-cell activation. Its binding to DPP4 induces T-cell proliferation and NF-kappa-B activation in a T-cell receptor/CD3-dependent manner. Recruits CTNNB1 to caveolar membranes and may regulate CTNNB1-mediated signaling through the Wnt pathway (By similarity). Negatively regulates TGFB1-mediated activation of SMAD2/3 by mediating the internalization of TGFBR1 from membrane rafts leading to its subsequent degradation.
Ubiquitinated. Undergo monoubiquitination and multi- and/or polyubiquitination. Monoubiquitination of N-terminal lysines promotes integration in a ternary complex with UBXN6 and VCP which promotes oligomeric CAV1 targeting to lysosomes for degradation.
The initiator methionine for isoform 2 is removed during or just after translation. The new N-terminal amino acid is then N-acetylated.
Phosphorylated at Tyr-14 by ABL1 in response to oxidative stress.
Golgi apparatus membrane>Peripheral membrane protein. Cell membrane>Peripheral membrane protein. Membrane>Caveola>Peripheral membrane protein. Membrane raft. Golgi apparatus>trans-Golgi network.
Note: Colocalized with DPP4 in membrane rafts. Potential hairpin-like structure in the membrane. Membrane protein of caveolae.
Skeletal muscle, liver, stomach, lung, kidney and heart (at protein level). Expressed in the brain.
Homooligomer. Interacts with GLIPR2. Interacts with NOSTRIN. Interacts with SNAP25 and STX1A (By similarity). Interacts (via the N-terminus) with DPP4; the interaction is direct. Interacts with CTNNB1, CDH1 and JUP. Interacts with PACSIN2; this interaction induces membrane tubulation (By similarity). Interacts with SLC7A9 (By similarity). Interacts with BMX and BTK. Interacts with TGFBR1. Interacts with CAVIN3 (via leucine-zipper domain) in a cholesterol-sensitive manner. Interacts with CAVIN1. Interacts with EHD2 in a cholesterol-dependent manner. Forms a ternary complex with UBXN6 and VCP; mediates CAV1 targeting to lysosomes for degradation. Interacts with ABCG1; this interaction regulates ABCG1-mediated cholesterol efflux.
(Microbial infection) Interacts with rotavirus A NSP4.
Belongs to the caveolin family.
Research Fields
· Cellular Processes > Transport and catabolism > Endocytosis. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cardiovascular diseases > Viral myocarditis.
References
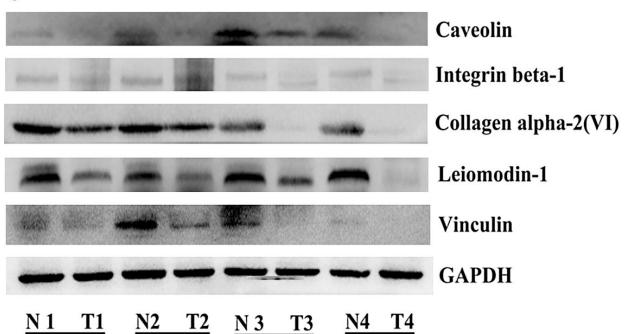
Application: WB Species: human Sample: ESCC tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.