DVL2 Antibody - #DF4454
| Product: | DVL2 Antibody |
| Catalog: | DF4454 |
| Description: | Rabbit polyclonal antibody to DVL2 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Zebrafish, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 90 KD; 79kD(Calculated). |
| Uniprot: | O14641 |
| RRID: | AB_2836809 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF4454, RRID:AB_2836809.
Fold/Unfold
Dishevelled 2 (homologous to Drosophila dsh); Dishevelled dsh homolog 2; dishevelled segment polarity protein 2; Dishevelled-2; Dishevelled2; DSH homolog 2; DVL 2; Dvl2; DVL2_HUMAN; Segment polarity protein dishevelled homolog DVL 2; Segment polarity protein dishevelled homolog DVL-2; Segment polarity protein dishevelled homolog DVL2;
Immunogens
- O14641 DVL2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAGSSTGGGGVGETKVIYHLDEEETPYLVKIPVPAERITLGDFKSVLQRPAGAKYFFKSMDQDFGVVKEEISDDNARLPCFNGRVVSWLVSSDNPQPEMAPPVHEPRAELAPPAPPLPPLPPERTSGIGDSRPPSFHPNVSSSHENLEPETETESVVSLRRERPRRRDSSEHGAGGHRTGGPSRLERHLAGYESSSTLMTSELESTSLGDSDEEDTMSRFSSSTEQSSASRLLKRHRRRRKQRPPRLERTSSFSSVTDSTMSLNIITVTLNMEKYNFLGISIVGQSNERGDGGIYIGSIMKGGAVAADGRIEPGDMLLQVNDMNFENMSNDDAVRVLRDIVHKPGPIVLTVAKCWDPSPQAYFTLPRNEPIQPIDPAAWVSHSAALTGTFPAYPGSSSMSTITSGSSLPDGCEGRGLSVHTDMASVTKAMAAPESGLEVRDRMWLKITIPNAFLGSDVVDWLYHHVEGFPERREARKYASGLLKAGLIRHTVNKITFSEQCYYVFGDLSGGCESYLVNLSLNDNDGSSGASDQDTLAPLPGATPWPLLPTFSYQYPAPHPYSPQPPPYHELSSYTYGGGSASSQHSEGSRSSGSTRSDGGAGRTGRPEERAPESKSGSGSESEPSSRGGSLRRGGEASGTSDGGPPPSRGSTGGAPNLRAHPGLHPYGPPPGMALPYNPMMVVMMPPPPPPVPPAVQPPGAPPVRDLGSVPPELTASRQSFHMAMGNPSEFFVDVM
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - O14641 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| Phosphorylation | Uniprot | ||
| Ubiquitination | Uniprot | ||
| T14 | Phosphorylation | Uniprot | |
| Y18 | Phosphorylation | P12931 (SRC) | Uniprot |
| Y27 | Phosphorylation | P12931 (SRC) | Uniprot |
| K30 | Ubiquitination | Uniprot | |
| T39 | Phosphorylation | Uniprot | |
| K44 | Ubiquitination | Uniprot | |
| K54 | Ubiquitination | Uniprot | |
| K58 | Ubiquitination | Uniprot | |
| S59 | Phosphorylation | Uniprot | |
| S135 | Phosphorylation | Uniprot | |
| S142 | Phosphorylation | Uniprot | |
| S143 | Phosphorylation | P49674 (CSNK1E) , P48730 (CSNK1D) | Uniprot |
| T151 | Phosphorylation | Uniprot | |
| T153 | Phosphorylation | Uniprot | |
| S155 | Phosphorylation | Uniprot | |
| S158 | Phosphorylation | Uniprot | |
| S169 | Phosphorylation | Uniprot | |
| S170 | Phosphorylation | Uniprot | |
| S195 | Phosphorylation | Uniprot | |
| S196 | Phosphorylation | Uniprot | |
| T206 | Phosphorylation | P53350 (PLK1) , PR:P53350 (hPLK1) | Uniprot |
| S211 | Phosphorylation | Uniprot | |
| S221 | Phosphorylation | Uniprot | |
| S222 | Phosphorylation | Uniprot | |
| T224 | Phosphorylation | P48730 (CSNK1D) , P49674 (CSNK1E) | Uniprot |
| S227 | Phosphorylation | Uniprot | |
| S228 | Phosphorylation | Uniprot | |
| S230 | Phosphorylation | Uniprot | |
| T250 | Phosphorylation | Uniprot | |
| S252 | Phosphorylation | Uniprot | |
| S254 | Phosphorylation | Uniprot | |
| S255 | Phosphorylation | Uniprot | |
| T257 | Phosphorylation | Uniprot | |
| Y275 | Phosphorylation | P12931 (SRC) | Uniprot |
| S286 | Phosphorylation | Uniprot | |
| Y295 | Phosphorylation | P12931 (SRC) | Uniprot |
| K301 | Ubiquitination | Uniprot | |
| K343 | Ubiquitination | Uniprot | |
| K353 | Ubiquitination | Uniprot | |
| S358 | Phosphorylation | Uniprot | |
| Y362 | Phosphorylation | Uniprot | |
| T364 | Phosphorylation | Uniprot | |
| S418 | Phosphorylation | Uniprot | |
| K428 | Ubiquitination | Uniprot | |
| Y463 | Phosphorylation | P12931 (SRC) | Uniprot |
| K477 | Ubiquitination | Uniprot | |
| Y478 | Phosphorylation | Uniprot | |
| K484 | Ubiquitination | Uniprot | |
| S591 | Phosphorylation | Uniprot | |
| S592 | Phosphorylation | Uniprot | |
| S594 | Phosphorylation | Uniprot | |
| T595 | Phosphorylation | Uniprot | |
| S597 | Phosphorylation | Uniprot | |
| S614 | Phosphorylation | Uniprot | |
| K615 | Ubiquitination | Uniprot | |
| S626 | Phosphorylation | Uniprot | |
| R627 | Methylation | Uniprot | |
| S630 | Phosphorylation | Uniprot | |
| R633 | Methylation | Uniprot | |
| R649 | Methylation | Uniprot | |
| S651 | Phosphorylation | Uniprot | |
| R659 | Methylation | Uniprot | |
| T715 | Phosphorylation | Uniprot | |
| S720 | Phosphorylation | Uniprot | |
| S729 | Phosphorylation | Uniprot |
Research Backgrounds
Plays a role in the signal transduction pathways mediated by multiple Wnt genes. Participates both in canonical and non-canonical Wnt signaling by binding to the cytoplasmic C-terminus of frizzled family members and transducing the Wnt signal to down-stream effectors. Promotes internalization and degradation of frizzled proteins upon Wnt signaling.
Phosphorylated by CSNK1D. WNT3A induces DVL2 phosphorylation by CSNK1E and MARK kinases.
Cell membrane>Peripheral membrane protein>Cytoplasmic side. Cytoplasm>Cytosol. Cytoplasmic vesicle. Nucleus.
Note: Localizes at the cell membrane upon interaction with frizzled family members and promotes their internalization. Localizes to cytoplasmic puncta (By similarity). Interaction with FOXK1 and FOXK2 induces nuclear translocation (PubMed:25805136).
Interacts through its PDZ domain with the C-terminal regions of VANGL1 and VANGL2. Interacts with Rac. Interacts with ARRB1; the interaction is enhanced by phosphorylation of DVL1 (By similarity). Can form large oligomers (via DIX domain). Interacts (via DIX domain) with DIXDC1 (via DIX domain). Interacts (via DEP domain) with AP2M1 and the AP-2 complex (By similarity). Interacts with DACT1 and FAM105B/otulin. Interacts with DCDC2. Interacts (when phosphorylated) with FOXK1 and FOXK2; the interaction induces DVL2 nuclear translocation. Interacts with MAPK15 (By similarity). Interacts with PKD1 (via extracellular domain). Interacts with LMBR1L (By similarity).
The DIX domain mediates homooligomerization.
Belongs to the DSH family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Notch signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Human papillomavirus infection.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Basal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Organismal Systems > Endocrine system > Melanogenesis.
References
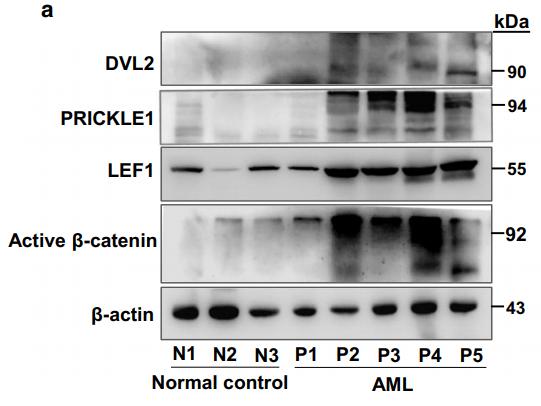
Application: WB Species: human Sample:
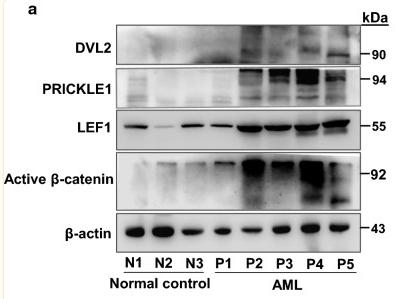
Application: WB Species: Human Sample:
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.