c-Myc Antibody - #AF0358
| Product: | c-Myc Antibody |
| Catalog: | AF0358 |
| Description: | Rabbit polyclonal antibody to c-Myc |
| Application: | WB IHC IF/ICC |
| Cited expt.: | WB, IHC, IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 49kDa; 49kD(Calculated). |
| Uniprot: | P01106 |
| RRID: | AB_2833523 |
Product Info
*The optimal dilutions should be determined by the end user. For optimal experimental results, antibody reuse is not recommended.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF0358, RRID:AB_2833523.
Fold/Unfold
AU016757; Avian myelocytomatosis viral oncogene homolog; bHLHe39; c Myc; Class E basic helix-loop-helix protein 39; MRTL; Myc; Myc protein; Myc proto oncogene protein; Myc proto-oncogene protein; myc-related translation/localization regulatory factor; MYC_HUMAN; Myc2; MYCC; Myelocytomatosis oncogene; Niard; Nird; Oncogene Myc; OTTHUMP00000158589; Proto-oncogene c-Myc; Protooncogene homologous to myelocytomatosis virus; RNCMYC; Transcription factor p64; Transcriptional regulator Myc-A; V-Myc avian myelocytomatosis viral oncogene homolog; v-myc myelocytomatosis viral oncogene homolog (avian);
Immunogens
A synthesized peptide derived from human c-Myc, corresponding to a region within N-terminal amino acids.
- P01106 MYC_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPLNVSFTNRNYDLDYDSVQPYFYCDEEENFYQQQQQSELQPPAPSEDIWKKFELLPTPPLSPSRRSGLCSPSYVAVTPFSLRGDNDGGGGSFSTADQLEMVTELLGGDMVNQSFICDPDDETFIKNIIIQDCMWSGFSAAAKLVSEKLASYQAARKDSGSPNPARGHSVCSTSSLYLQDLSAAASECIDPSVVFPYPLNDSSSPKSCASQDSSAFSPSSDSLLSSTESSPQGSPEPLVLHEETPPTTSSDSEEEQEDEEEIDVVSVEKRQAPGKRSESGSPSAGGHSKPPHSPLVLKRCHVSTHQHNYAAPPSTRKDYPAAKRVKLDSVRVLRQISNNRKCTSPRSSDTEENVKRRTHNVLERQRRNELKRSFFALRDQIPELENNEKAPKVVILKKATAYILSVQAEEQKLISEEDLLRKRREQLKHKLEQLRNSCA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
Research Backgrounds
Transcription factor that binds DNA in a non-specific manner, yet also specifically recognizes the core sequence 5'-CAC[GA]TG-3'. Activates the transcription of growth-related genes. Binds to the VEGFA promoter, promoting VEGFA production and subsequent sprouting angiogenesis. Regulator of somatic reprogramming, controls self-renewal of embryonic stem cells. Functions with TAF6L to activate target gene expression through RNA polymerase II pause release (By similarity).
Phosphorylated by PRKDC. Phosphorylation at Ser-329 by PIM2 leads to the stabilization of MYC (By similarity). Phosphorylation at Ser-62 by CDK2 prevents Ras-induced senescence. Phosphorylated at Ser-62 by DYRK2; this primes the protein for subsequent phosphorylation by GSK3B at Thr-58. Phosphorylation at Thr-58 and Ser-62 by GSK3 is required for ubiquitination and degradation by the proteasome.
Ubiquitinated by the SCF(FBXW7) complex when phosphorylated at Thr-58 and Ser-62, leading to its degradation by the proteasome. In the nucleoplasm, ubiquitination is counteracted by USP28, which interacts with isoform 1 of FBXW7 (FBW7alpha), leading to its deubiquitination and preventing degradation. In the nucleolus, however, ubiquitination is not counteracted by USP28 but by USP36, due to the lack of interaction between isoform 3 of FBXW7 (FBW7gamma) and USP28, explaining the selective MYC degradation in the nucleolus. Also polyubiquitinated by the DCX(TRUSS) complex. Ubiquitinated by TRIM6 in a phosphorylation-independent manner (By similarity).
Nucleus>Nucleoplasm. Nucleus>Nucleolus.
Research Fields
· Cellular Processes > Cell growth and death > Cell cycle. (View pathway)
· Cellular Processes > Cell growth and death > Cellular senescence. (View pathway)
· Cellular Processes > Cellular community - eukaryotes > Signaling pathways regulating pluripotency of stem cells. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > PI3K-Akt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Wnt signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > TGF-beta signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Hippo signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Jak-STAT signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Viral > Hepatitis B.
· Human Diseases > Infectious diseases: Viral > HTLV-I infection.
· Human Diseases > Infectious diseases: Viral > Epstein-Barr virus infection.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > Transcriptional misregulation in cancer.
· Human Diseases > Cancers: Overview > Proteoglycans in cancer.
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Colorectal cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Endometrial cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Thyroid cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Bladder cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Acute myeloid leukemia. (View pathway)
· Human Diseases > Cancers: Specific types > Small cell lung cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Breast cancer. (View pathway)
· Human Diseases > Cancers: Specific types > Hepatocellular carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Gastric cancer. (View pathway)
· Human Diseases > Cancers: Overview > Central carbon metabolism in cancer. (View pathway)
· Organismal Systems > Endocrine system > Thyroid hormone signaling pathway. (View pathway)
References
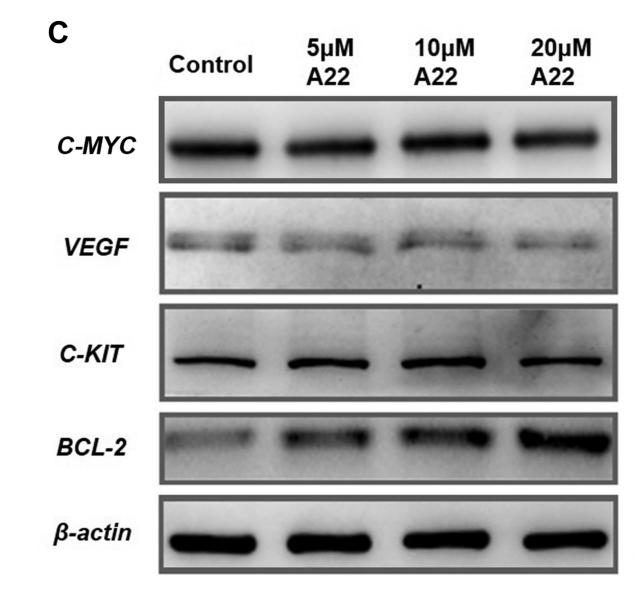
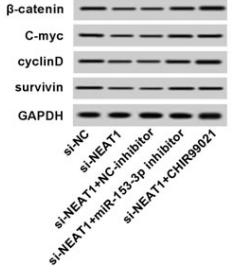
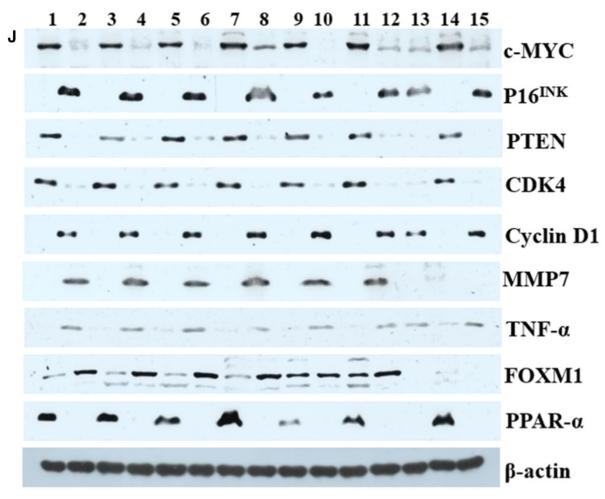
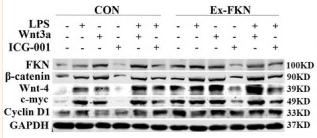
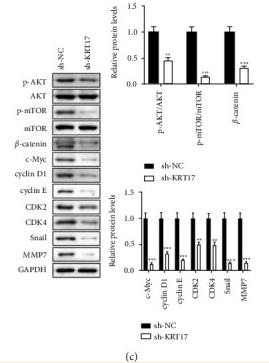
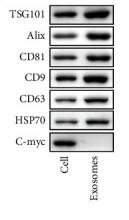
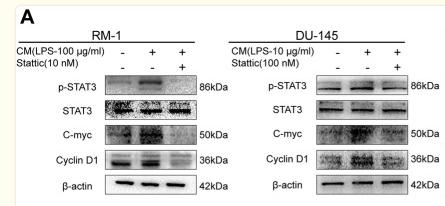
Application: WB Species: human Sample: HepG2
Application: WB Species: Mouse Sample: tumor tissue
Application: IHC Species: Mouse Sample: tumor tissue
Application: WB Species: Human Sample: KYSE30 and TE-1 cells
Application: WB Species: Human Sample: pancreatic cancer cells
Application: WB Species: mouse Sample: J774A. 1 cells
Application: WB Species: Mice Sample: J774A.1 cells
Application: WB Species: Human Sample: HCC cells
Application: WB Species: human Sample: NSE-overexpressing H446 cells
Application: WB Species: Human Sample: SCLC cells
Application: WB Species: Human Sample: SCLC cells
Application: WB Species: Human Sample: sclc cells
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.