Crk p38 Antibody - #AF6323
| Product: | Crk p38 Antibody |
| Catalog: | AF6323 |
| Description: | Rabbit polyclonal antibody to Crk p38 |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat, Monkey |
| Prediction: | Pig, Bovine, Horse, Sheep, Dog, Chicken, Xenopus |
| Mol.Wt.: | 40kDa; 34kD(Calculated). |
| Uniprot: | P46108 |
| RRID: | AB_2835181 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6323, RRID:AB_2835181.
Fold/Unfold
Adapter molecule crk; Avian sarcoma virus CT10 (v crk) oncogene homolog; CRK; CRK isoform 2; CRK isoform II; CRK_HUMAN; CRKII; FLJ38130; OTTHUMP00000115366; OTTHUMP00000198330; p38; Proto oncogene C crk; Proto-oncogene C-crk; v crk avian sarcoma virus CT10 oncogene homolog; v crk sarcoma virus CT10 oncogene homolog; v crk sarcoma virus CT10 oncogene homolog (avian);
Immunogens
- P46108 CRK_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MAGNFDSEERSSWYWGRLSRQEAVALLQGQRHGVFLVRDSSTSPGDYVLSVSENSRVSHYIINSSGPRPPVPPSPAQPPPGVSPSRLRIGDQEFDSLPALLEFYKIHYLDTTTLIEPVSRSRQGSGVILRQEEAEYVRALFDFNGNDEEDLPFKKGDILRIRDKPEEQWWNAEDSEGKRGMIPVPYVEKYRPASASVSALIGGNQEGSHPQPLGGPEPGPYAQPSVNTPLPNLQNGPIYARVIQKRVPNAYDKTALALEVGELVKVTKINVSGQWEGECNGKRGHFPFTHVRLLDQQNPDEDFS
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P46108 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| A2 | Acetylation | Uniprot | |
| S40 | Phosphorylation | Uniprot | |
| S41 | Phosphorylation | Q13153 (PAK1) | Uniprot |
| T42 | Phosphorylation | Uniprot | |
| S43 | Phosphorylation | Uniprot | |
| Y47 | Phosphorylation | Uniprot | |
| S74 | Phosphorylation | Uniprot | |
| S83 | Phosphorylation | Uniprot | |
| S85 | Phosphorylation | Uniprot | |
| S96 | Phosphorylation | Uniprot | |
| Y108 | Phosphorylation | Uniprot | |
| T111 | Phosphorylation | Uniprot | |
| T112 | Phosphorylation | Uniprot | |
| T113 | Phosphorylation | Uniprot | |
| S121 | Phosphorylation | Uniprot | |
| R122 | Methylation | Uniprot | |
| S125 | Phosphorylation | Uniprot | |
| Y136 | Phosphorylation | Uniprot | |
| K154 | Acetylation | Uniprot | |
| S175 | Phosphorylation | Uniprot | |
| Y190 | Phosphorylation | Uniprot | |
| S194 | Phosphorylation | Uniprot | |
| S196 | Phosphorylation | Uniprot | |
| S198 | Phosphorylation | Uniprot | |
| S208 | Phosphorylation | Uniprot | |
| Y221 | Phosphorylation | P00519 (ABL1) , P00533 (EGFR) , A0A173G4P4 (Abl fusion) , P08069 (IGF1R) , P42684 (ABL2) | Uniprot |
| S225 | Phosphorylation | Uniprot | |
| T228 | Phosphorylation | Uniprot | |
| Y239 | Phosphorylation | P00519 (ABL1) | Uniprot |
| Y251 | Phosphorylation | P00519 (ABL1) | Uniprot |
| K253 | Acetylation | Uniprot | |
| T254 | Phosphorylation | Uniprot | |
| S304 | Phosphorylation | Uniprot |
Research Backgrounds
Regulates cell adhesion, spreading and migration. Mediates attachment-induced MAPK8 activation, membrane ruffling and cell motility in a Rac-dependent manner. Involved in phagocytosis of apoptotic cells and cell motility via its interaction with DOCK1 and DOCK4. May regulate the EFNA5-EPHA3 signaling.
Phosphorylation of Crk-II (40 kDa) gives rise to a 42 kDa form. Isoform Crk-II is phosphorylated by KIT.
Phosphorylated on Tyr-221 upon cell adhesion. Results in the negative regulation of the association with SH2- and SH3-binding partners, possibly by the formation of an intramolecular interaction of phosphorylated Tyr-221 with the SH2 domain. This leads finally to the down-regulation of the Crk signaling pathway.
Proline isomerization at Pro-237 by PPIA acts as a switch between two conformations: an autoinhibitory conformation in the cis form, where the tandem SH3 domains interact intramolecularly, and an activated conformation in the trans form.
Cytoplasm. Cell membrane.
Note: Translocated to the plasma membrane upon cell adhesion.
Interacts with ABL1, C3G, DOCK3, DOCK5, MAP4K1, MAPK8 and SOS via its first SH3 domain. Interacts (via SH2 domain) with BCAR1, CBL, CBLB, PXN, IRS4 and GAB1 upon stimulus-induced tyrosine phosphorylation. Interacts (via SH2 domain) with several tyrosine-phosphorylated growth factor receptors such as EGFR and INSR. Interacts with FLT1 (tyrosine-phosphorylated). Interacts with DOCK1 and DOCK4. Interacts with SHB. Interacts with PEAK1. Interacts with FASLG. Isoform Crk-II interacts with KIT. Interacts with EPHA3; upon activation of EPHA3 by the ligand EFNA5 and EPHA3 tyrosine kinase activity-dependent. Interacts with EPHA3 (phosphorylated); mediates EFNA5-EPHA3 signaling through RHOA GTPase activation. Interacts with FLT4 (tyrosine-phosphorylated). Isoform Crk-II (via SH2 domain) interacts with PDGFRA (tyrosine phosphorylated) and PDGFRB (tyrosine phosphorylated). Part of a collagen stimulated complex involved in cell migration composed of CDC42, CRK, TNK2 and p130cas/BCAR1. Interacts (via SH2 domain) with the 'Tyr-9' phosphorylated form of PDPK1. Interacts with CBLC. Found in a complex with ABL1, ABL2, CRK and UNC119; leading to the inhibition of CRK phosphorylation by ABL kinases.
The C-terminal SH3 domain function as a negative modulator for transformation and the N-terminal SH3 domain appears to function as a positive regulator for transformation.
The SH2 domain mediates interaction with tyrosine phosphorylated proteins. Mediates interaction with SHB.
Belongs to the CRK family.
Research Fields
· Cellular Processes > Cellular community - eukaryotes > Focal adhesion. (View pathway)
· Cellular Processes > Cell motility > Regulation of actin cytoskeleton. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > ErbB signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > Rap1 signaling pathway. (View pathway)
· Human Diseases > Infectious diseases: Bacterial > Bacterial invasion of epithelial cells.
· Human Diseases > Infectious diseases: Bacterial > Shigellosis.
· Human Diseases > Cancers: Overview > Pathways in cancer. (View pathway)
· Human Diseases > Cancers: Overview > MicroRNAs in cancer.
· Human Diseases > Cancers: Specific types > Renal cell carcinoma. (View pathway)
· Human Diseases > Cancers: Specific types > Chronic myeloid leukemia. (View pathway)
· Organismal Systems > Immune system > Chemokine signaling pathway. (View pathway)
· Organismal Systems > Immune system > Fc gamma R-mediated phagocytosis. (View pathway)
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Insulin signaling pathway. (View pathway)
References
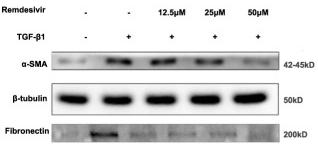
Application: WB Species: Mice Sample: lung tissues
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.