AhR Antibody - #AF6278
| Product: | AhR Antibody |
| Catalog: | AF6278 |
| Description: | Rabbit polyclonal antibody to AhR |
| Application: | WB IHC IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 96kDa; 96kD(Calculated). |
| Uniprot: | P35869 |
| RRID: | AB_2835131 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# AF6278, RRID:AB_2835131.
Fold/Unfold
Ah receptor; AhR; AHR_HUMAN; Aromatic hydrocarbon receptor; Aryl hydrocarbon receptor; Aryl hydrocarbon receptor precursor; bHLHe76; Class E basic helix loop helix protein 76; Class E basic helix-loop-helix protein 76; HGNC:348;
Immunogens
Expressed in all tissues tested including blood, brain, heart, kidney, liver, lung, pancreas and skeletal muscle. Expressed in retinal photoreceptors (PubMed:29726989).
- P35869 AHR_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MNSSSANITYASRKRRKPVQKTVKPIPAEGIKSNPSKRHRDRLNTELDRLASLLPFPQDVINKLDKLSVLRLSVSYLRAKSFFDVALKSSPTERNGGQDNCRAANFREGLNLQEGEFLLQALNGFVLVVTTDALVFYASSTIQDYLGFQQSDVIHQSVYELIHTEDRAEFQRQLHWALNPSQCTESGQGIEEATGLPQTVVCYNPDQIPPENSPLMERCFICRLRCLLDNSSGFLAMNFQGKLKYLHGQKKKGKDGSILPPQLALFAIATPLQPPSILEIRTKNFIFRTKHKLDFTPIGCDAKGRIVLGYTEAELCTRGSGYQFIHAADMLYCAESHIRMIKTGESGMIVFRLLTKNNRWTWVQSNARLLYKNGRPDYIIVTQRPLTDEEGTEHLRKRNTKLPFMFTTGEAVLYEATNPFPAIMDPLPLRTKNGTSGKDSATTSTLSKDSLNPSSLLAAMMQQDESIYLYPASSTSSTAPFENNFFNESMNECRNWQDNTAPMGNDTILKHEQIDQPQDVNSFAGGHPGLFQDSKNSDLYSIMKNLGIDFEDIRHMQNEKFFRNDFSGEVDFRDIDLTDEILTYVQDSLSKSPFIPSDYQQQQSLALNSSCMVQEHLHLEQQQQHHQKQVVVEPQQQLCQKMKHMQVNGMFENWNSNQFVPFNCPQQDPQQYNVFTDLHGISQEFPYKSEMDSMPYTQNFISCNQPVLPQHSKCTELDYPMGSFEPSPYPTTSSLEDFVTCLQLPENQKHGLNPQSAIITPQTCYAGAVSMYQCQPEPQHTHVGQMQYNPVLPGQQAFLNKFQNGVLNETYPAELNNINNTQTTTHLQPLHHPSEARPFPDLTSSGFL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - P35869 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| M1 | Acetylation | Uniprot | |
| S12 | Phosphorylation | P17252 (PRKCA) | Uniprot |
| K24 | Acetylation | Uniprot | |
| K24 | Sumoylation | Uniprot | |
| K24 | Ubiquitination | Uniprot | |
| K32 | Ubiquitination | Uniprot | |
| S36 | Phosphorylation | P17252 (PRKCA) | Uniprot |
| T45 | Phosphorylation | Uniprot | |
| K63 | Ubiquitination | Uniprot | |
| S68 | Phosphorylation | Uniprot | |
| S75 | Phosphorylation | Uniprot | |
| Y76 | Phosphorylation | Uniprot | |
| S81 | Phosphorylation | Uniprot | |
| K88 | Ubiquitination | Uniprot | |
| S232 | Phosphorylation | Uniprot | |
| K244 | Ubiquitination | Uniprot | |
| K254 | Sumoylation | Uniprot | |
| K292 | Ubiquitination | Uniprot | |
| Y322 | Phosphorylation | Uniprot | |
| T355 | Phosphorylation | Uniprot | |
| Y378 | Phosphorylation | Uniprot | |
| K438 | Ubiquitination | Uniprot | |
| K510 | Ubiquitination | Uniprot | |
| K535 | Ubiquitination | Uniprot | |
| Y540 | Phosphorylation | Uniprot | |
| K544 | Ubiquitination | Uniprot | |
| K560 | Ubiquitination | Uniprot | |
| K641 | Ubiquitination | Uniprot |
Research Backgrounds
Ligand-activated transcriptional activator. Binds to the XRE promoter region of genes it activates. Activates the expression of multiple phase I and II xenobiotic chemical metabolizing enzyme genes (such as the CYP1A1 gene). Mediates biochemical and toxic effects of halogenated aromatic hydrocarbons. Involved in cell-cycle regulation. Likely to play an important role in the development and maturation of many tissues. Regulates the circadian clock by inhibiting the basal and circadian expression of the core circadian component PER1. Inhibits PER1 by repressing the CLOCK-ARNTL/BMAL1 heterodimer mediated transcriptional activation of PER1. The heterodimer ARNT:AHR binds to core DNA sequence 5'-TGCGTG-3' within the dioxin response element (DRE) of target gene promoters and activates their transcription.
Mono-ADP-ribosylated, leading to inhibit transcription activator activity of AHR.
Cytoplasm. Nucleus.
Note: Initially cytoplasmic; upon binding with ligand and interaction with a HSP90, it translocates to the nucleus.
Expressed in all tissues tested including blood, brain, heart, kidney, liver, lung, pancreas and skeletal muscle. Expressed in retinal photoreceptors.
Homodimer (By similarity). Binds MYBBP1A (By similarity). Efficient DNA binding requires dimerization with another bHLH protein. Interacts with coactivators including SRC-1, RIP140 and NOCA7, and with the corepressor SMRT. Interacts with NEDD8 and IVNS1ABP. Interacts with ARNTL/BMAL1. Interacts with HSP90AB1 (By similarity). Interacts with ARNT; the heterodimer ARNT:AHR binds to core DNA sequence 5'-TGCGTG-3' within the dioxin response element (DRE) of target gene promoters and activates their transcription. Interacts with TIPARP; leading to mono-ADP-ribosylation of AHR and subsequent inhibition of AHR.
The PAS 1 domain is essential for dimerization and also required for AHR:ARNT heterodimerization.
Research Fields
· Organismal Systems > Immune system > Th17 cell differentiation. (View pathway)
References
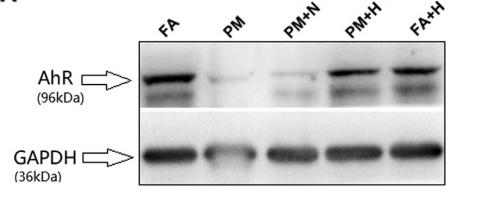
Application: WB Species: rat Sample: lung
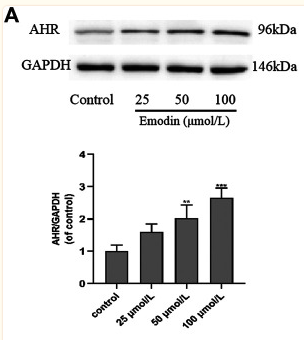
Application: WB Species: Human Sample: MCF-7 cells
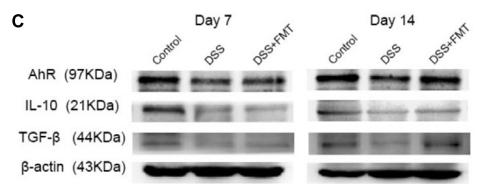
Application: WB Species: mouse Sample: colon
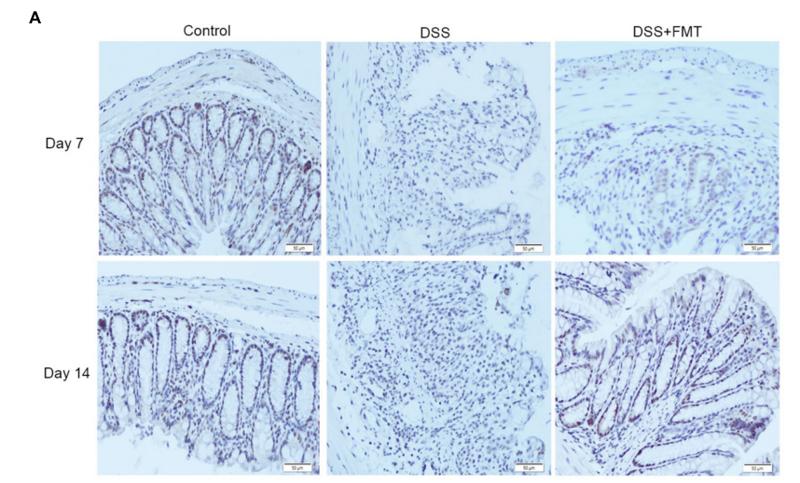
Application: IHC Species: mouse Sample: colon
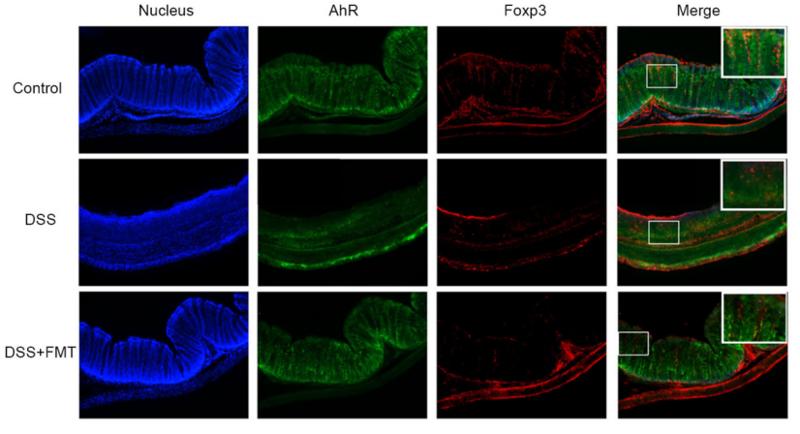
Application: IF/ICC Species: mouse Sample: colon
Application: WB Species: Human Sample: Periodontal ligament fibroblasts (PDLFs)
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.