Phospho-RSK1/2/3/4 (Ser221/Ser227/Ser218/Ser232) Antibody - #DF8743
| Product: | Phospho-RSK1/2/3/4 (Ser221/Ser227/Ser218/Ser232) Antibody |
| Catalog: | DF8743 |
| Description: | Rabbit polyclonal antibody to Phospho-RSK1/2/3/4 (Ser221/Ser227/Ser218/Ser232) |
| Application: | WB IF/ICC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Xenopus |
| Mol.Wt.: | 82 kDa; 83kD,84kD(Calculated). |
| Uniprot: | Q15418 | P51812 | Q15349 | Q9UK32 |
| RRID: | AB_2841947 |
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF8743, RRID:AB_2841947.
Fold/Unfold
90 kDa ribosomal protein S6 kinase 1; dJ590P13.1 (ribosomal protein S6 kinase, 90kD, polypeptide 1; dJ590P13.1; EC 2.7.11.1; HU 1; HU1; KS6A1_HUMAN; MAP kinase activated protein kinase 1a; MAP kinase-activated protein kinase 1a; MAPK-activated protein kinase 1a; MAPKAP kinase 1a; MAPKAPK-1a; MAPKAPK1A; MGC79981; Mitogen-activated protein kinase-activated protein kinase 1A; OTTHUMP00000004113; p90 RSK1; p90-RSK 1; p90rsk; p90RSK1; p90S6K; pp90RSK1; Ribosomal protein S6 kinase 90kD 1; Ribosomal protein S6 kinase 90kD polypeptide 1; Ribosomal protein S6 kinase 90kDa polypeptide 1; Ribosomal protein S6 kinase alpha 1; Ribosomal protein S6 kinase alpha-1; Ribosomal protein S6 kinase polypeptide 1; Ribosomal S6 kinase 1; RPS6K1 alpha; rps6ka; Rps6ka1; RSK 1; RSK 1 p90; RSK; RSK-1; RSK1; S6K alpha 1; S6K-alpha-1; 90 kDa ribosomal protein S6 kinase 3; CLS; HU 3; HU2; HU3; Insulin stimulated protein kinase 1; Insulin-stimulated protein kinase 1; ISPK-1; ISPK1; KS6A3_HUMAN; MAP kinase activated protein kinase 1b; MAP kinase-activated protein kinase 1b; MAPK activated protein kinase 1b; MAPK-activated protein kinase 1b; MAPKAP kinase 1b; MAPKAPK 1b; MAPKAPK-1b; MAPKAPK1B; Mental retardation, X linked 19; MRX19; OTTHUMP00000023036; p90 RSK2; p90 RSK3; p90-RSK 3; p90RSK3; pp90RSK2; Ribosomal protein S6 kinase 90kDa polypeptide 3; Ribosomal protein S6 kinase alpha 3; Ribosomal protein S6 kinase alpha-3; Ribosomal protein s6 kinase ii alpha 2; Ribosomal S6 kinase 2; Rps6ka3; RSK; RSK-2; RSK2; S6 kinase 2; S6K alpha3; S6K-alpha-3; 90 kDa ribosomal protein S6 kinase 2; HU 2; KS6A2_HUMAN; MAP kinase activated protein kinase 1c; MAP kinase-activated protein kinase 1c; MAPK-activated protein kinase 1c; MAPKAP kinase 1c; MAPKAPK-1c; MAPKAPK1C; Mitogen.activated protein kinase-activated protein kinase 1C; p90 RSK3; p90-RSK 2; p90RSK2; pp90RSK3; Ribosomal protein S6 kinase alpha-2; ribosomal protein S6 kinase, 90kDa, polypeptide 2; Ribosomal S6 kinase 3; RPS6KA2; RSK 3; RSK; RSK-3; S6K alpha; S6K alpha 2; S6K-alpha-2; 90 kDa ribosomal protein S6 kinase 6; KS6A6_HUMAN; p90 RSK 6; p90-RSK 6; p90RSK6; pp90RSK4; Ribosomal protein S6 kinase 90kDa polypeptide 6; Ribosomal protein S6 kinase alpha 6; Ribosomal protein S6 kinase alpha-6; Ribosomal S6 kinase 4; rps6ka6; RSK 4; RSK-4; S6K-alpha-6;
Immunogens
Q15418(KS6A1_HUMAN) >>Visit HPA database.
P51812(KS6A3_HUMAN) >>Visit HPA database.
Expressed in many tissues, highest levels in skeletal muscle.
Q15349 KS6A2_HUMAN:Widely expressed with higher expression in lung, skeletal muscle, brain, uterus, ovary, thyroid and prostate.
- Q15418 KS6A1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPLAQLKEPWPLMELVPLDPENGQTSGEEAGLQPSKDEGVLKEISITHHVKAGSEKADPSHFELLKVLGQGSFGKVFLVRKVTRPDSGHLYAMKVLKKATLKVRDRVRTKMERDILADVNHPFVVKLHYAFQTEGKLYLILDFLRGGDLFTRLSKEVMFTEEDVKFYLAELALGLDHLHSLGIIYRDLKPENILLDEEGHIKLTDFGLSKEAIDHEKKAYSFCGTVEYMAPEVVNRQGHSHSADWWSYGVLMFEMLTGSLPFQGKDRKETMTLILKAKLGMPQFLSTEAQSLLRALFKRNPANRLGSGPDGAEEIKRHVFYSTIDWNKLYRREIKPPFKPAVAQPDDTFYFDTEFTSRTPKDSPGIPPSAGAHQLFRGFSFVATGLMEDDGKPRAPQAPLHSVVQQLHGKNLVFSDGYVVKETIGVGSYSECKRCVHKATNMEYAVKVIDKSKRDPSEEIEILLRYGQHPNIITLKDVYDDGKHVYLVTELMRGGELLDKILRQKFFSEREASFVLHTIGKTVEYLHSQGVVHRDLKPSNILYVDESGNPECLRICDFGFAKQLRAENGLLMTPCYTANFVAPEVLKRQGYDEGCDIWSLGILLYTMLAGYTPFANGPSDTPEEILTRIGSGKFTLSGGNWNTVSETAKDLVSKMLHVDPHQRLTAKQVLQHPWVTQKDKLPQSQLSHQDLQLVKGAMAATYSALNSSKPTPQLKPIESSILAQRRVRKLPSTTL
- P51812 KS6A3_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MPLAQLADPWQKMAVESPSDSAENGQQIMDEPMGEEEINPQTEEVSIKEIAITHHVKEGHEKADPSQFELLKVLGQGSFGKVFLVKKISGSDARQLYAMKVLKKATLKVRDRVRTKMERDILVEVNHPFIVKLHYAFQTEGKLYLILDFLRGGDLFTRLSKEVMFTEEDVKFYLAELALALDHLHSLGIIYRDLKPENILLDEEGHIKLTDFGLSKESIDHEKKAYSFCGTVEYMAPEVVNRRGHTQSADWWSFGVLMFEMLTGTLPFQGKDRKETMTMILKAKLGMPQFLSPEAQSLLRMLFKRNPANRLGAGPDGVEEIKRHSFFSTIDWNKLYRREIHPPFKPATGRPEDTFYFDPEFTAKTPKDSPGIPPSANAHQLFRGFSFVAITSDDESQAMQTVGVHSIVQQLHRNSIQFTDGYEVKEDIGVGSYSVCKRCIHKATNMEFAVKIIDKSKRDPTEEIEILLRYGQHPNIITLKDVYDDGKYVYVVTELMKGGELLDKILRQKFFSEREASAVLFTITKTVEYLHAQGVVHRDLKPSNILYVDESGNPESIRICDFGFAKQLRAENGLLMTPCYTANFVAPEVLKRQGYDAACDIWSLGVLLYTMLTGYTPFANGPDDTPEEILARIGSGKFSLSGGYWNSVSDTAKDLVSKMLHVDPHQRLTAALVLRHPWIVHWDQLPQYQLNRQDAPHLVKGAMAATYSALNRNQSPVLEPVGRSTLAQRRGIKKITSTAL
- Q15349 KS6A2_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MDLSMKKFAVRRFFSVYLRRKSRSKSSSLSRLEEEGVVKEIDISHHVKEGFEKADPSQFELLKVLGQGSYGKVFLVRKVKGSDAGQLYAMKVLKKATLKVRDRVRSKMERDILAEVNHPFIVKLHYAFQTEGKLYLILDFLRGGDLFTRLSKEVMFTEEDVKFYLAELALALDHLHSLGIIYRDLKPENILLDEEGHIKITDFGLSKEAIDHDKRAYSFCGTIEYMAPEVVNRRGHTQSADWWSFGVLMFEMLTGSLPFQGKDRKETMALILKAKLGMPQFLSGEAQSLLRALFKRNPCNRLGAGIDGVEEIKRHPFFVTIDWNTLYRKEIKPPFKPAVGRPEDTFHFDPEFTARTPTDSPGVPPSANAHHLFRGFSFVASSLIQEPSQQDLHKVPVHPIVQQLHGNNIHFTDGYEIKEDIGVGSYSVCKRCVHKATDTEYAVKIIDKSKRDPSEEIEILLRYGQHPNIITLKDVYDDGKFVYLVMELMRGGELLDRILRQRYFSEREASDVLCTITKTMDYLHSQGVVHRDLKPSNILYRDESGSPESIRVCDFGFAKQLRAGNGLLMTPCYTANFVAPEVLKRQGYDAACDIWSLGILLYTMLAGFTPFANGPDDTPEEILARIGSGKYALSGGNWDSISDAAKDVVSKMLHVDPHQRLTAMQVLKHPWVVNREYLSPNQLSRQDVHLVKGAMAATYFALNRTPQAPRLEPVLSSNLAQRRGMKRLTSTRL
- Q9UK32 KS6A6_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MLPFAPQDEPWDREMEVFSGGGASSGEVNGLKMVDEPMEEGEADSCHDEGVVKEIPITHHVKEGYEKADPAQFELLKVLGQGSFGKVFLVRKKTGPDAGQLYAMKVLKKASLKVRDRVRTKMERDILVEVNHPFIVKLHYAFQTEGKLYLILDFLRGGDVFTRLSKEVLFTEEDVKFYLAELALALDHLHQLGIVYRDLKPENILLDEIGHIKLTDFGLSKESVDQEKKAYSFCGTVEYMAPEVVNRRGHSQSADWWSYGVLMFEMLTGTLPFQGKDRNETMNMILKAKLGMPQFLSAEAQSLLRMLFKRNPANRLGSEGVEEIKRHLFFANIDWDKLYKREVQPPFKPASGKPDDTFCFDPEFTAKTPKDSPGLPASANAHQLFKGFSFVATSIAEEYKITPITSANVLPIVQINGNAAQFGEVYELKEDIGVGSYSVCKRCIHATTNMEFAVKIIDKSKRDPSEEIEILMRYGQHPNIITLKDVFDDGRYVYLVTDLMKGGELLDRILKQKCFSEREASDILYVISKTVDYLHCQGVVHRDLKPSNILYMDESASADSIRICDFGFAKQLRGENGLLLTPCYTANFVAPEVLMQQGYDAACDIWSLGVLFYTMLAGYTPFANGPNDTPEEILLRIGNGKFSLSGGNWDNISDGAKDLLSHMLHMDPHQRYTAEQILKHSWITHRDQLPNDQPKRNDVSHVVKGAMVATYSALTHKTFQPVLEPVAASSLAQRRSMKKRTSTGL
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - Q15418/P51812/Q15349/Q9UK32 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S26 | Phosphorylation | Uniprot | |
| S27 | Phosphorylation | Uniprot | |
| S28 | Phosphorylation | Uniprot | |
| S30 | Phosphorylation | Uniprot | |
| K91 | Acetylation | Uniprot | |
| K94 | Acetylation | Uniprot | |
| S106 | Phosphorylation | Uniprot | |
| Y135 | Phosphorylation | Uniprot | |
| K186 | Ubiquitination | Uniprot | |
| S206 | Phosphorylation | Uniprot | |
| Y217 | Phosphorylation | Uniprot | |
| S218 | Phosphorylation | O15530 (PDPK1) , Q15349 (RPS6KA2) | Uniprot |
| T222 | Phosphorylation | Uniprot | |
| K295 | Acetylation | Uniprot | |
| T356 | Phosphorylation | Uniprot | |
| S360 | Phosphorylation | Uniprot | |
| S377 | Phosphorylation | Uniprot | |
| S381 | Phosphorylation | Uniprot | |
| S388 | Phosphorylation | Uniprot | |
| Y426 | Phosphorylation | Uniprot | |
| K430 | Ubiquitination | Uniprot | |
| K450 | Ubiquitination | Uniprot | |
| Y463 | Phosphorylation | Uniprot | |
| T471 | Phosphorylation | Uniprot | |
| K473 | Ubiquitination | Uniprot | |
| S546 | Phosphorylation | Uniprot | |
| T570 | Phosphorylation | Q15349 (RPS6KA2) | Uniprot |
| Y573 | Phosphorylation | Uniprot | |
| T574 | Phosphorylation | Uniprot | |
| T618 | Phosphorylation | Uniprot | |
| S628 | Phosphorylation | Uniprot | |
| K630 | Ubiquitination | Uniprot | |
| S650 | Phosphorylation | Uniprot | |
| T662 | Phosphorylation | Uniprot | |
| S730 | Phosphorylation | Uniprot |
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S17 | Phosphorylation | Uniprot | |
| S19 | Phosphorylation | Uniprot | |
| K57 | Ubiquitination | Uniprot | |
| K62 | Ubiquitination | Uniprot | |
| K72 | Ubiquitination | Uniprot | |
| S78 | Phosphorylation | Uniprot | |
| K81 | Acetylation | Uniprot | |
| K100 | Acetylation | Uniprot | |
| K103 | Acetylation | Uniprot | |
| Y144 | Phosphorylation | Uniprot | |
| K195 | Ubiquitination | Uniprot | |
| S215 | Phosphorylation | Uniprot | |
| K216 | Ubiquitination | Uniprot | |
| Y226 | Phosphorylation | Uniprot | |
| S227 | Phosphorylation | P51812 (RPS6KA3) , P27361 (MAPK3) , O15530 (PDPK1) | Uniprot |
| T231 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| Y234 | Phosphorylation | Uniprot | |
| K274 | Ubiquitination | Uniprot | |
| T278 | Phosphorylation | Uniprot | |
| K322 | Ubiquitination | Uniprot | |
| K345 | Ubiquitination | Uniprot | |
| Y356 | Phosphorylation | Uniprot | |
| T362 | Phosphorylation | Uniprot | |
| K364 | Ubiquitination | Uniprot | |
| T365 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| S369 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| S375 | Phosphorylation | Uniprot | |
| S386 | Phosphorylation | P28482 (MAPK1) , P51812 (RPS6KA3) , P49137 (MAPKAPK2) , O15530 (PDPK1) | Uniprot |
| T391 | Phosphorylation | Uniprot | |
| S392 | Phosphorylation | Uniprot | |
| S396 | Phosphorylation | Uniprot | |
| S415 | Phosphorylation | Uniprot | |
| Y433 | Phosphorylation | Uniprot | |
| K437 | Ubiquitination | Uniprot | |
| Y470 | Phosphorylation | P12931 (SRC) , P06241 (FYN) | Uniprot |
| T478 | Phosphorylation | Uniprot | |
| K480 | Ubiquitination | Uniprot | |
| Y483 | Phosphorylation | P06241 (FYN) , P12931 (SRC) | Uniprot |
| Y488 | Phosphorylation | P12931 (SRC) , P06241 (FYN) , P28482 (MAPK1) , P22607 (FGFR3) | Uniprot |
| K504 | Ubiquitination | Uniprot | |
| K509 | Ubiquitination | Uniprot | |
| S517 | Phosphorylation | Uniprot | |
| Y529 | Phosphorylation | P12931 (SRC) , P28482 (MAPK1) , P22607 (FGFR3) , P06241 (FYN) | Uniprot |
| K541 | Ubiquitination | Uniprot | |
| S556 | Phosphorylation | Uniprot | |
| K566 | Ubiquitination | Uniprot | |
| T573 | Phosphorylation | Uniprot | |
| T577 | Phosphorylation | P27361 (MAPK3) | Uniprot |
| Y580 | Phosphorylation | P06241 (FYN) , P12931 (SRC) | Uniprot |
| T581 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| K591 | Ubiquitination | Uniprot | |
| T625 | Phosphorylation | Uniprot | |
| Y644 | Phosphorylation | P12931 (SRC) , P06241 (FYN) | Uniprot |
| K658 | Ubiquitination | Uniprot | |
| Y688 | Phosphorylation | Uniprot | |
| T706 | Phosphorylation | Uniprot | |
| Y707 | Phosphorylation | P06241 (FYN) , P35916 (FLT4) , P11362 (FGFR1) , P22607 (FGFR3) , P12931 (SRC) | Uniprot |
| S708 | Phosphorylation | Uniprot | |
| S715 | Phosphorylation | P51812 (RPS6KA3) | Uniprot |
| R723 | Methylation | Uniprot | |
| S737 | Phosphorylation | Uniprot |
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S45 | Phosphorylation | Uniprot | |
| T47 | Phosphorylation | Uniprot | |
| K51 | Ubiquitination | Uniprot | |
| S54 | Phosphorylation | Uniprot | |
| K56 | Ubiquitination | Uniprot | |
| K66 | Ubiquitination | Uniprot | |
| S72 | Phosphorylation | Uniprot | |
| K75 | Acetylation | Uniprot | |
| K75 | Ubiquitination | Uniprot | |
| K81 | Ubiquitination | Uniprot | |
| Y91 | Phosphorylation | Uniprot | |
| K94 | Ubiquitination | Uniprot | |
| Y138 | Phosphorylation | Uniprot | |
| S154 | Phosphorylation | Uniprot | |
| K189 | Ubiquitination | Uniprot | |
| S209 | Phosphorylation | Uniprot | |
| K210 | Ubiquitination | Uniprot | |
| Y220 | Phosphorylation | Uniprot | |
| S221 | Phosphorylation | Q15418 (RPS6KA1) , O15530 (PDPK1) | Uniprot |
| T225 | Phosphorylation | Uniprot | |
| Y228 | Phosphorylation | Uniprot | |
| K278 | Ubiquitination | Uniprot | |
| S307 | Phosphorylation | Uniprot | |
| K316 | Ubiquitination | Uniprot | |
| T359 | Phosphorylation | Uniprot | |
| S363 | Phosphorylation | Uniprot | |
| S369 | Phosphorylation | Uniprot | |
| S380 | Phosphorylation | P49137 (MAPKAPK2) , P41279 (MAP3K8) , Q13237 (PRKG2) , Q15418 (RPS6KA1) | Uniprot |
| T384 | Phosphorylation | Uniprot | |
| K392 | Ubiquitination | Uniprot | |
| S402 | Phosphorylation | Uniprot | |
| K410 | Ubiquitination | Uniprot | |
| K421 | Ubiquitination | Uniprot | |
| K433 | Ubiquitination | Uniprot | |
| K438 | Ubiquitination | Uniprot | |
| K453 | Ubiquitination | Uniprot | |
| Y466 | Phosphorylation | Uniprot | |
| T474 | Phosphorylation | Uniprot | |
| K476 | Ubiquitination | Uniprot | |
| Y486 | Phosphorylation | Uniprot | |
| T489 | Phosphorylation | Uniprot | |
| K500 | Ubiquitination | Uniprot | |
| K505 | Ubiquitination | Uniprot | |
| S513 | Phosphorylation | Uniprot | |
| T522 | Phosphorylation | Uniprot | |
| K537 | Ubiquitination | Uniprot | |
| K562 | Ubiquitination | Uniprot | |
| T573 | Phosphorylation | P28482 (MAPK1) | Uniprot |
| Y576 | Phosphorylation | Uniprot | |
| T577 | Phosphorylation | Uniprot | |
| K587 | Ubiquitination | Uniprot | |
| K633 | Ubiquitination | Uniprot | |
| S637 | Phosphorylation | Uniprot | |
| K649 | Ubiquitination | Uniprot | |
| K654 | Ubiquitination | Uniprot | |
| K667 | Ubiquitination | Uniprot | |
| K678 | Ubiquitination | Uniprot | |
| K680 | Ubiquitination | Uniprot | |
| S684 | Phosphorylation | Uniprot | |
| S687 | Phosphorylation | Uniprot | |
| T701 | Phosphorylation | Uniprot | |
| Y702 | Phosphorylation | Uniprot | |
| S703 | Phosphorylation | Uniprot | |
| K709 | Ubiquitination | Uniprot | |
| S732 | Phosphorylation | Uniprot | |
| T733 | Phosphorylation | Uniprot | |
| T734 | Phosphorylation | Uniprot |
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| S83 | Phosphorylation | Uniprot | |
| K86 | Acetylation | Uniprot | |
| K86 | Ubiquitination | Uniprot | |
| T94 | Phosphorylation | Uniprot | |
| K105 | Acetylation | Uniprot | |
| K108 | Acetylation | Uniprot | |
| Y149 | Phosphorylation | Uniprot | |
| S220 | Phosphorylation | Uniprot | |
| Y231 | Phosphorylation | Uniprot | |
| S232 | Phosphorylation | Uniprot | |
| T236 | Phosphorylation | Uniprot | |
| Y239 | Phosphorylation | Uniprot | |
| K348 | Acetylation | Uniprot | |
| K367 | Acetylation | Uniprot | |
| S372 | Phosphorylation | Uniprot | |
| S378 | Phosphorylation | Uniprot | |
| K386 | Acetylation | Uniprot | |
| S389 | Phosphorylation | Uniprot | |
| Y437 | Phosphorylation | Uniprot | |
| K441 | Ubiquitination | Uniprot | |
| Y474 | Phosphorylation | Uniprot | |
| T482 | Phosphorylation | Uniprot | |
| S521 | Phosphorylation | Uniprot | |
| Y525 | Phosphorylation | Uniprot | |
| S547 | Phosphorylation | Uniprot | |
| S555 | Phosphorylation | Uniprot | |
| K570 | Ubiquitination | Uniprot | |
| T581 | Phosphorylation | Uniprot | |
| T710 | Phosphorylation | Uniprot | |
| Y711 | Phosphorylation | Uniprot | |
| T715 | Phosphorylation | Uniprot | |
| T718 | Phosphorylation | Uniprot | |
| S730 | Phosphorylation | Uniprot | |
| S736 | Phosphorylation | Uniprot |
PTMs - Q15418/P51812/Q15349/Q9UK32 As Enzyme
| Substrate | Site | Source |
|---|---|---|
| P16220 (CREB1) | S133 | Uniprot |
| P17676 (CEBPB) | S321 | Uniprot |
| P25963 (NFKBIA) | S32 | Uniprot |
| P32004-2 (L1CAM) | S1152 | Uniprot |
| Q15349-1 (RPS6KA2) | S218 | Uniprot |
| Q15349-1 (RPS6KA2) | T222 | Uniprot |
| Q15349 (RPS6KA2) | T570 | Uniprot |
| Q92934 (BAD) | S75 | Uniprot |
| Q92934 (BAD) | S118 | Uniprot |
| Substrate | Site | Source |
|---|---|---|
| O14649 (KCNK3) | S393 | Uniprot |
| O60825 (PFKFB2) | S466 | Uniprot |
| P03372 (ESR1) | S167 | Uniprot |
| P04637 (TP53) | S15 | Uniprot |
| P04792 (HSPB1) | S78 | Uniprot |
| P04792 (HSPB1) | S82 | Uniprot |
| P05114 (HMGN1) | S7 | Uniprot |
| P05114 (HMGN1) | S21 | Uniprot |
| P05114 (HMGN1) | S25 | Uniprot |
| P05198 (EIF2S1) | S52 | Uniprot |
| P05204 (HMGN2) | S25 | Uniprot |
| P05204 (HMGN2) | S29 | Uniprot |
| P05783 (KRT18) | S53 | Uniprot |
| P07101 (TH) | S40 | Uniprot |
| P07101-4 (TH) | S44 | Uniprot |
| P07101-2 (TH) | S67 | Uniprot |
| P07101 (TH) | S71 | Uniprot |
| P10636-6 (MAPT) | T154 | Uniprot |
| P10636-8 (MAPT) | T212 | Uniprot |
| P11362 (FGFR1) | S789 | Uniprot |
| P13796 (LCP1) | S5 | Uniprot |
| P16220 (CREB1) | S133 | Uniprot |
| P17676 (CEBPB) | S321 | Uniprot |
| P18846 (ATF1) | S63 | Uniprot |
| P18848 (ATF4) | S245 | Uniprot |
| P19525 (EIF2AK2) | T451 | Uniprot |
| P22736-1 (NR4A1) | S351 | Uniprot |
| P23588 (EIF4B) | S422 | Uniprot |
| P25963 (NFKBIA) | S32 | Uniprot |
| P29317 (EPHA2) | S897 | Uniprot |
| P33778 (HIST1H2BB) | S33 | Uniprot |
| P40763 (STAT3) | S727 | Uniprot |
| P46527 (CDKN1B) | T198 | Uniprot |
| P48764 (SLC9A3) | S663 | Uniprot |
| P49815 (TSC2) | S1798 | Uniprot |
| P49840 (GSK3A) | S21 | Uniprot |
| P49841 (GSK3B) | S9 | Uniprot |
| P51812 (RPS6KA3) | S227 | Uniprot |
| P51812 (RPS6KA3) | T231 | Uniprot |
| P51812 (RPS6KA3) | T365 | Uniprot |
| P51812 (RPS6KA3) | S369 | Uniprot |
| P51812 (RPS6KA3) | S386 | Uniprot |
| P51812 (RPS6KA3) | T581 | Uniprot |
| P51812 (RPS6KA3) | S715 | Uniprot |
| P53355 (DAPK1) | S289 | Uniprot |
| P62753 (RPS6) | S235 | Uniprot |
| P62753 (RPS6) | S236 | Uniprot |
| P67809 (YBX1) | S102 | Uniprot |
| P68431 (HIST1H3J) | S11 | Uniprot |
| P84243 (H3F3B) | S11 | Uniprot |
| Q04206 (RELA) | S536 | Uniprot |
| Q06413 (MEF2C) | S192 | Uniprot |
| Q14790 (CASP8) | T263 | Uniprot |
| Q14934 (NFATC4) | S281 | Uniprot |
| Q14934 (NFATC4) | S285 | Uniprot |
| Q14934 (NFATC4) | S289 | Uniprot |
| Q14934 (NFATC4) | S344 | Uniprot |
| Q14934 (NFATC4) | S676 | Uniprot |
| Q15831-1 (STK11) | S428 | Uniprot |
| Q16695 (HIST3H3) | S11 | Uniprot |
| Q16821 (PPP1R3A) | S46 | Uniprot |
| Q16821 (PPP1R3A) | S65 | Uniprot |
| Q8IX03 (WWC1) | T929 | Uniprot |
| Q8IX03 (WWC1) | S947 | Uniprot |
| Q8N122 (RPTOR) | S719 | Uniprot |
| Q8N122 (RPTOR) | S721 | Uniprot |
| Q8N122 (RPTOR) | S722 | Uniprot |
| Q92934 (BAD) | S75 | Uniprot |
| Q92934 (BAD) | S99 | Uniprot |
| Q96RK0 (CIC) | S173 | Uniprot |
| Q99683 (MAP3K5) | S83 | Uniprot |
| Q99683 (MAP3K5) | T1109 | Uniprot |
| Q99683 (MAP3K5) | T1326 | Uniprot |
| Q9BSI4 (TINF2) | S295 | Uniprot |
| Q9BSI4 (TINF2) | S330 | Uniprot |
| Q9H6Z4 (RANBP3) | S126 | Uniprot |
| Q9NYV6 (RRN3) | S649 | Uniprot |
| Substrate | Site | Source |
|---|---|---|
| O00418 (EEF2K) | S366 | Uniprot |
| O14727 (APAF1) | S268 | Uniprot |
| O14727 (APAF1) | S357 | Uniprot |
| O14757 (CHEK1) | S280 | Uniprot |
| O60343 (TBC1D4) | S341 | Uniprot |
| O60343 (TBC1D4) | S751 | Uniprot |
| O75030-9 (MITF) | S409 | Uniprot |
| P01100 (FOS) | S362 | Uniprot |
| P03372-1 (ESR1) | S118 | Uniprot |
| P03372-1 (ESR1) | S167 | Uniprot |
| P11831 (SRF) | S103 | Uniprot |
| P13796 (LCP1) | S5 | Uniprot |
| P16144 (ITGB4) | S1364 | Uniprot |
| P16220-1 (CREB1) | S133 | Uniprot |
| P17676 (CEBPB) | T235 | Uniprot |
| P17676-1 (CEBPB) | T266 | Uniprot |
| P19634 (SLC9A1) | S703 | Uniprot |
| P21333 (FLNA) | S2152 | Uniprot |
| P22736-1 (NR4A1) | S351 | Uniprot |
| P23588 (EIF4B) | S422 | Uniprot |
| P25963 (NFKBIA) | S32 | Uniprot |
| P25963 (NFKBIA) | S36 | Uniprot |
| P29317 (EPHA2) | S897 | Uniprot |
| P30304 (CDC25A) | S293 | Uniprot |
| P30304 (CDC25A) | S295 | Uniprot |
| P30305 (CDC25B) | S353 | Uniprot |
| P30305 (CDC25B) | T355 | Uniprot |
| P30307 (CDC25C) | S247 | Uniprot |
| P32004-1 (L1CAM) | S1152 | Uniprot |
| P42345 (MTOR) | T2446 | Uniprot |
| P42345 (MTOR) | S2448 | Uniprot |
| P43354-1 (NR4A2) | S347 | Uniprot |
| P46527 (CDKN1B) | T198 | Uniprot |
| P49427 (CDC34) | T162 | Uniprot |
| P49815 (TSC2) | S1798 | Uniprot |
| P49841 (GSK3B) | S9 | Uniprot |
| P50549 (ETV1) | S191 | Uniprot |
| P50549 (ETV1) | S216 | Uniprot |
| P50549-1 (ETV1) | S334 | Uniprot |
| P50552 (VASP) | T278 | Uniprot |
| P53355 (DAPK1) | S289 | Uniprot |
| P60983 (GMFB) | T27 | Uniprot |
| P62753 (RPS6) | S235 | Uniprot |
| P62753 (RPS6) | S236 | Uniprot |
| P62753 (RPS6) | S240 | Uniprot |
| P62753 (RPS6) | S244 | Uniprot |
| P67809 (YBX1) | S102 | Uniprot |
| P78371 (CCT2) | S260 | Uniprot |
| P84243 (H3F3B) | S11 | Uniprot |
| Q04206 (RELA) | S536 | Uniprot |
| Q05195 (MXD1) | S145 | Uniprot |
| Q07889 (SOS1) | S1134 | Uniprot |
| Q07889 (SOS1) | S1161 | Uniprot |
| Q13164 (MAPK7) | S496 | Uniprot |
| Q15418 (RPS6KA1) | S221 | Uniprot |
| Q15418-1 (RPS6KA1) | T225 | Uniprot |
| Q15418-2 (RPS6KA1) | T234 | Uniprot |
| Q15418-1 (RPS6KA1) | S380 | Uniprot |
| Q15418-2 (RPS6KA1) | S389 | Uniprot |
| Q15418-1 (RPS6KA1) | T577 | Uniprot |
| Q15418-2 (RPS6KA1) | T586 | Uniprot |
| Q15653 (NFKBIB) | S19 | Uniprot |
| Q15653 (NFKBIB) | S23 | Uniprot |
| Q15831 (STK11) | S428 | Uniprot |
| Q16821-1 (PPP1R3A) | S46 | Uniprot |
| Q16875 (PFKFB3) | S461 | Uniprot |
| Q8IX03 (WWC1) | T929 | Uniprot |
| Q8IX03 (WWC1) | S947 | Uniprot |
| Q8N122 (RPTOR) | S719 | Uniprot |
| Q8N122 (RPTOR) | S721 | Uniprot |
| Q8N122 (RPTOR) | S722 | Uniprot |
| Q8TB45 (DEPTOR) | S286 | Uniprot |
| Q8TB45 (DEPTOR) | S287 | Uniprot |
| Q8TB45 (DEPTOR) | S291 | Uniprot |
| Q92570 (NR4A3) | S376 | Uniprot |
| Q92570-3 (NR4A3) | S387 | Uniprot |
| Q92882 (OSTF1) | S202 | Uniprot |
| Q92934 (BAD) | S75 | Uniprot |
| Q92934 (BAD) | S99 | Uniprot |
| Q92934 (BAD) | S118 | Uniprot |
| Q92934 (BAD) | S153 | Uniprot |
| Q96RK0 (CIC) | S173 | Uniprot |
| Q9H6Z4 (RANBP3) | S126 | Uniprot |
| Q9HC62 (SENP2) | T369 | Uniprot |
| Q9UBP6-1 (METTL1) | S27 | Uniprot |
| Q9UN36 (NDRG2) | S332 | Uniprot |
| Q9UN36 (NDRG2) | S350 | Uniprot |
| Q9Y2V2 (CARHSP1) | S52 | Uniprot |
Research Backgrounds
Serine/threonine-protein kinase that acts downstream of ERK (MAPK1/ERK2 and MAPK3/ERK1) signaling and mediates mitogenic and stress-induced activation of the transcription factors CREB1, ETV1/ER81 and NR4A1/NUR77, regulates translation through RPS6 and EIF4B phosphorylation, and mediates cellular proliferation, survival, and differentiation by modulating mTOR signaling and repressing pro-apoptotic function of BAD and DAPK1. In fibroblast, is required for EGF-stimulated phosphorylation of CREB1, which results in the subsequent transcriptional activation of several immediate-early genes. In response to mitogenic stimulation (EGF and PMA), phosphorylates and activates NR4A1/NUR77 and ETV1/ER81 transcription factors and the cofactor CREBBP. Upon insulin-derived signal, acts indirectly on the transcription regulation of several genes by phosphorylating GSK3B at 'Ser-9' and inhibiting its activity. Phosphorylates RPS6 in response to serum or EGF via an mTOR-independent mechanism and promotes translation initiation by facilitating assembly of the pre-initiation complex. In response to insulin, phosphorylates EIF4B, enhancing EIF4B affinity for the EIF3 complex and stimulating cap-dependent translation. Is involved in the mTOR nutrient-sensing pathway by directly phosphorylating TSC2 at 'Ser-1798', which potently inhibits TSC2 ability to suppress mTOR signaling, and mediates phosphorylation of RPTOR, which regulates mTORC1 activity and may promote rapamycin-sensitive signaling independently of the PI3K/AKT pathway. Mediates cell survival by phosphorylating the pro-apoptotic proteins BAD and DAPK1 and suppressing their pro-apoptotic function. Promotes the survival of hepatic stellate cells by phosphorylating CEBPB in response to the hepatotoxin carbon tetrachloride (CCl4). Mediates induction of hepatocyte prolifration by TGFA through phosphorylation of CEBPB (By similarity). Is involved in cell cycle regulation by phosphorylating the CDK inhibitor CDKN1B, which promotes CDKN1B association with 14-3-3 proteins and prevents its translocation to the nucleus and inhibition of G1 progression. Phosphorylates EPHA2 at 'Ser-897', the RPS6KA-EPHA2 signaling pathway controls cell migration.
Activated by phosphorylation at Ser-221 by PDPK1. Autophosphorylated on Ser-380, as part of the activation process. May be phosphorylated at Thr-359 and Ser-363 by MAPK1/ERK2 and MAPK3/ERK1.
N-terminal myristoylation results in an activated kinase in the absence of added growth factors.
Nucleus. Cytoplasm.
Forms a complex with either MAPK1/ERK2 or MAPK3/ERK1 in quiescent cells. Transiently dissociates following mitogenic stimulation. Interacts with ETV1/ER81 and FGFR1.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. S6 kinase subfamily.
Serine/threonine-protein kinase that acts downstream of ERK (MAPK1/ERK2 and MAPK3/ERK1) signaling and mediates mitogenic and stress-induced activation of the transcription factors CREB1, ETV1/ER81 and NR4A1/NUR77, regulates translation through RPS6 and EIF4B phosphorylation, and mediates cellular proliferation, survival, and differentiation by modulating mTOR signaling and repressing pro-apoptotic function of BAD and DAPK1. In fibroblast, is required for EGF-stimulated phosphorylation of CREB1 and histone H3 at 'Ser-10', which results in the subsequent transcriptional activation of several immediate-early genes. In response to mitogenic stimulation (EGF and PMA), phosphorylates and activates NR4A1/NUR77 and ETV1/ER81 transcription factors and the cofactor CREBBP. Upon insulin-derived signal, acts indirectly on the transcription regulation of several genes by phosphorylating GSK3B at 'Ser-9' and inhibiting its activity. Phosphorylates RPS6 in response to serum or EGF via an mTOR-independent mechanism and promotes translation initiation by facilitating assembly of the preinitiation complex. In response to insulin, phosphorylates EIF4B, enhancing EIF4B affinity for the EIF3 complex and stimulating cap-dependent translation. Is involved in the mTOR nutrient-sensing pathway by directly phosphorylating TSC2 at 'Ser-1798', which potently inhibits TSC2 ability to suppress mTOR signaling, and mediates phosphorylation of RPTOR, which regulates mTORC1 activity and may promote rapamycin-sensitive signaling independently of the PI3K/AKT pathway. Mediates cell survival by phosphorylating the pro-apoptotic proteins BAD and DAPK1 and suppressing their pro-apoptotic function. Promotes the survival of hepatic stellate cells by phosphorylating CEBPB in response to the hepatotoxin carbon tetrachloride (CCl4). Is involved in cell cycle regulation by phosphorylating the CDK inhibitor CDKN1B, which promotes CDKN1B association with 14-3-3 proteins and prevents its translocation to the nucleus and inhibition of G1 progression. In LPS-stimulated dendritic cells, is involved in TLR4-induced macropinocytosis, and in myeloma cells, acts as effector of FGFR3-mediated transformation signaling, after direct phosphorylation at Tyr-529 by FGFR3. Negatively regulates EGF-induced MAPK1/3 phosphorylation via phosphorylation of SOS1. Phosphorylates SOS1 at 'Ser-1134' and 'Ser-1161' that create YWHAB and YWHAE binding sites and which contribute to the negative regulation of MAPK1/3 phosphorylation (By similarity). Phosphorylates EPHA2 at 'Ser-897', the RPS6KA-EPHA2 signaling pathway controls cell migration.
Activated by phosphorylation at Ser-227 by PDPK1. Autophosphorylated on Ser-386, as part of the activation process. May be phosphorylated at Thr-365 and Ser-369 by MAPK1/ERK2 and MAPK3/ERK1. Can also be activated via phosphorylation at Ser-386 by MAPKAPK2.
N-terminal myristoylation results in an activated kinase in the absence of added growth factors.
Nucleus. Cytoplasm.
Expressed in many tissues, highest levels in skeletal muscle.
Forms a complex with either MAPK1/ERK2 or MAPK3/ERK1 in quiescent cells. Transiently dissociates following mitogenic stimulation (By similarity). Interacts with NFATC4, ETV1/ER81 and FGFR1.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. S6 kinase subfamily.
Serine/threonine-protein kinase that acts downstream of ERK (MAPK1/ERK2 and MAPK3/ERK1) signaling and mediates mitogenic and stress-induced activation of transcription factors, regulates translation, and mediates cellular proliferation, survival, and differentiation. May function as tumor suppressor in epithelial ovarian cancer cells.
Activated by phosphorylation at Ser-218 by PDPK1. Autophosphorylated on Ser-377, as part of the activation process. May be phosphorylated at Thr-356 and Ser-360 by MAPK1/ERK2 and MAPK3/ERK1 (By similarity).
N-terminal myristoylation results in an activated kinase in the absence of added growth factors.
Nucleus. Cytoplasm.
Widely expressed with higher expression in lung, skeletal muscle, brain, uterus, ovary, thyroid and prostate.
Forms a complex with either MAPK1/ERK2 or MAPK3/ERK1 in quiescent cells. Transiently dissociates following mitogenic stimulation (By similarity). Interacts with FBXO5; cooperate to induce the metaphase arrest of early blastomeres; increases and stabilizes interaction of FBXO5 with CDC20 (By similarity).
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. S6 kinase subfamily.
Constitutively active serine/threonine-protein kinase that exhibits growth-factor-independent kinase activity and that may participate in p53/TP53-dependent cell growth arrest signaling and play an inhibitory role during embryogenesis.
Phosphorylated at Ser-232, Ser-372, and Ser-389 in serum-starved cells.
Cytoplasm>Cytosol. Nucleus.
Note: Predominantly cytosolic.
Forms a complex with MAPK3/ERK1 but not with MAPK9 or MAPK14 in serum-starved cells.
Belongs to the protein kinase superfamily. AGC Ser/Thr protein kinase family. S6 kinase subfamily.
Research Fields
· Cellular Processes > Cell growth and death > Oocyte meiosis. (View pathway)
· Environmental Information Processing > Signal transduction > MAPK signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Human Diseases > Endocrine and metabolic diseases > Insulin resistance.
· Organismal Systems > Nervous system > Long-term potentiation.
· Organismal Systems > Nervous system > Neurotrophin signaling pathway. (View pathway)
· Organismal Systems > Endocrine system > Progesterone-mediated oocyte maturation.
References
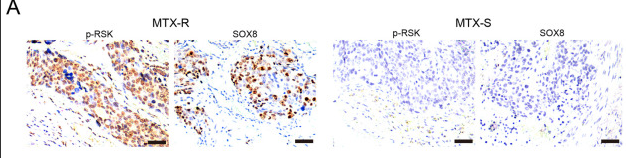
Application: IHC Species: Human Sample: GTN
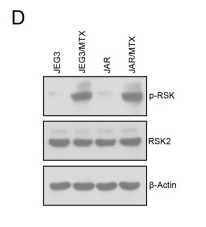
Application: WB Species: Human Sample: JAR/MTX and JAR cell
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.