ULK1 Antibody - #DF7588
| Product: | ULK1 Antibody |
| Catalog: | DF7588 |
| Description: | Rabbit polyclonal antibody to ULK1 |
| Application: | WB IHC |
| Reactivity: | Human, Mouse, Rat |
| Prediction: | Pig, Bovine, Horse, Sheep, Rabbit, Dog, Chicken, Xenopus |
| Mol.Wt.: | 120~150 kDa; 113kD(Calculated). |
| Uniprot: | O75385 |
| RRID: | AB_2841079 |
Related Downloads
Protocols
Product Info
*The optimal dilutions should be determined by the end user.
*Tips:
WB: For western blot detection of denatured protein samples. IHC: For immunohistochemical detection of paraffin sections (IHC-p) or frozen sections (IHC-f) of tissue samples. IF/ICC: For immunofluorescence detection of cell samples. ELISA(peptide): For ELISA detection of antigenic peptide.
Cite Format: Affinity Biosciences Cat# DF7588, RRID:AB_2841079.
Fold/Unfold
ATG 1; ATG1; ATG1 autophagy related 1 homolog; ATG1A; Autophagy related protein 1 homolog; Autophagy-related protein 1 homolog; FLJ38455; FLJ46475; hATG1; KIAA0722; Serine/threonine protein kinase ULK1; Serine/threonine protein kinase Unc51.1; Serine/threonine-protein kinase ULK1; ULK 1; ULK1; ULK1_HUMAN; Unc 51 (C. elegans) like kinase 1; UNC 51; Unc 51 like kinase 1; Unc-51 like kinase 1 (C. elegans); Unc-51-like kinase 1; UNC51; UNC51, C. elegans, homolog of; Unc51.1;
Immunogens
Ubiquitously expressed. Detected in the following adult tissues: skeletal muscle, heart, pancreas, brain, placenta, liver, kidney, and lung.
- O75385 ULK1_HUMAN:
- Protein BLAST With
- NCBI/
- ExPASy/
- Uniprot
MEPGRGGTETVGKFEFSRKDLIGHGAFAVVFKGRHREKHDLEVAVKCINKKNLAKSQTLLGKEIKILKELKHENIVALYDFQEMANSVYLVMEYCNGGDLADYLHAMRTLSEDTIRLFLQQIAGAMRLLHSKGIIHRDLKPQNILLSNPAGRRANPNSIRVKIADFGFARYLQSNMMAATLCGSPMYMAPEVIMSQHYDGKADLWSIGTIVYQCLTGKAPFQASSPQDLRLFYEKNKTLVPTIPRETSAPLRQLLLALLQRNHKDRMDFDEFFHHPFLDASPSVRKSPPVPVPSYPSSGSGSSSSSSSTSHLASPPSLGEMQQLQKTLASPADTAGFLHSSRDSGGSKDSSCDTDDFVMVPAQFPGDLVAEAPSAKPPPDSLMCSGSSLVASAGLESHGRTPSPSPPCSSSPSPSGRAGPFSSSRCGASVPIPVPTQVQNYQRIERNLQSPTQFQTPRSSAIRRSGSTSPLGFARASPSPPAHAEHGGVLARKMSLGGGRPYTPSPQVGTIPERPGWSGTPSPQGAEMRGGRSPRPGSSAPEHSPRTSGLGCRLHSAPNLSDLHVVRPKLPKPPTDPLGAVFSPPQASPPQPSHGLQSCRNLRGSPKLPDFLQRNPLPPILGSPTKAVPSFDFPKTPSSQNLLALLARQGVVMTPPRNRTLPDLSEVGPFHGQPLGPGLRPGEDPKGPFGRSFSTSRLTDLLLKAAFGTQAPDPGSTESLQEKPMEIAPSAGFGGSLHPGARAGGTSSPSPVVFTVGSPPSGSTPPQGPRTRMFSAGPTGSASSSARHLVPGPCSEAPAPELPAPGHGCSFADPITANLEGAVTFEAPDLPEETLMEQEHTEILRGLRFTLLFVQHVLEIAALKGSASEAAGGPEYQLQESVVADQISLLSREWGFAEQLVLYLKVAELLSSGLQSAIDQIRAGKLCLSSTVKQVVRRLNELYKASVVSCQGLSLRLQRFFLDKQRLLDRIHSITAERLIFSHAVQMVQSAALDEMFQHREGCVPRYHKALLLLEGLQHMLSDQADIENVTKCKLCIERRLSALLTGICA
Predictions
Score>80(red) has high confidence and is suggested to be used for WB detection. *The prediction model is mainly based on the alignment of immunogen sequences, the results are for reference only, not as the basis of quality assurance.
High(score>80) Medium(80>score>50) Low(score<50) No confidence
PTMs - O75385 As Substrate
| Site | PTM Type | Enzyme | Source |
|---|---|---|---|
| K13 | Ubiquitination | Uniprot | |
| K46 | Ubiquitination | Uniprot | |
| K55 | Ubiquitination | Uniprot | |
| K62 | Ubiquitination | Uniprot | |
| S111 | Phosphorylation | Uniprot | |
| S131 | Phosphorylation | Uniprot | |
| K140 | Ubiquitination | Uniprot | |
| S158 | Phosphorylation | Uniprot | |
| K162 | Acetylation | Uniprot | |
| K162 | Ubiquitination | Uniprot | |
| T180 | Phosphorylation | O75385 (ULK1) | Uniprot |
| S225 | Phosphorylation | Uniprot | |
| K235 | Ubiquitination | Uniprot | |
| K237 | Ubiquitination | Uniprot | |
| S281 | Phosphorylation | Uniprot | |
| S317 | Phosphorylation | Q13131 (PRKAA1) , P54646 (PRKAA2) | Uniprot |
| S330 | Phosphorylation | Uniprot | |
| S403 | Phosphorylation | Uniprot | |
| S405 | Phosphorylation | Uniprot | |
| S409 | Phosphorylation | Uniprot | |
| S415 | Phosphorylation | Uniprot | |
| S429 | Phosphorylation | Uniprot | |
| S450 | Phosphorylation | Uniprot | |
| T456 | Phosphorylation | Uniprot | |
| S459 | Phosphorylation | Uniprot | |
| S460 | Phosphorylation | Uniprot | |
| S465 | Phosphorylation | Uniprot | |
| S467 | Phosphorylation | Uniprot | |
| T468 | Phosphorylation | Uniprot | |
| S469 | Phosphorylation | Uniprot | |
| S477 | Phosphorylation | Uniprot | |
| S479 | Phosphorylation | Uniprot | |
| S495 | Phosphorylation | Uniprot | |
| S522 | Phosphorylation | Uniprot | |
| R529 | Methylation | Uniprot | |
| S533 | Phosphorylation | Uniprot | |
| S538 | Phosphorylation | Uniprot | |
| S539 | Phosphorylation | Uniprot | |
| S544 | Phosphorylation | Uniprot | |
| S556 | Phosphorylation | P54646 (PRKAA2) | Uniprot |
| S561 | Phosphorylation | Uniprot | |
| T575 | Phosphorylation | Uniprot | |
| S583 | Phosphorylation | Uniprot | |
| S588 | Phosphorylation | Uniprot | |
| S593 | Phosphorylation | Uniprot | |
| S598 | Phosphorylation | Uniprot | |
| S605 | Phosphorylation | Uniprot | |
| K607 | Acetylation | Uniprot | |
| S623 | Phosphorylation | Uniprot | |
| T625 | Phosphorylation | Uniprot | |
| S630 | Phosphorylation | Uniprot | |
| T636 | Phosphorylation | Uniprot | |
| S638 | Phosphorylation | P42345 (MTOR) , P54646 (PRKAA2) , Q13131 (PRKAA1) | Uniprot |
| S639 | Phosphorylation | Uniprot | |
| T654 | Phosphorylation | Uniprot | |
| T660 | Phosphorylation | Uniprot | |
| S665 | Phosphorylation | Uniprot | |
| S694 | Phosphorylation | Uniprot | |
| T695 | Phosphorylation | Uniprot | |
| T699 | Phosphorylation | Uniprot | |
| T709 | Phosphorylation | Uniprot | |
| S716 | Phosphorylation | Uniprot | |
| T717 | Phosphorylation | Uniprot | |
| S719 | Phosphorylation | Uniprot | |
| S748 | Phosphorylation | Uniprot | |
| S758 | Phosphorylation | P42345 (MTOR) | Uniprot |
| S761 | Phosphorylation | Uniprot | |
| T764 | Phosphorylation | Uniprot | |
| S775 | Phosphorylation | Uniprot | |
| S781 | Phosphorylation | Uniprot | |
| S949 | Phosphorylation | Uniprot | |
| S954 | Phosphorylation | Uniprot | |
| S1042 | Phosphorylation | Uniprot | |
| T1046 | Phosphorylation | Uniprot |
PTMs - O75385 As Enzyme
| Substrate | Site | Source |
|---|---|---|
| A2RUS2 (DENND3) | S472 | Uniprot |
| A2RUS2 (DENND3) | S490 | Uniprot |
| O00560 (SDCBP) | S6 | Uniprot |
| O75143 (ATG13) | S355 | Uniprot |
| O75385 (ULK1) | T180 | Uniprot |
| Q13131 (PRKAA1) | S360 | Uniprot |
| Q13131 (PRKAA1) | T368 | Uniprot |
| Q13131 (PRKAA1) | S397 | Uniprot |
| Q13131 (PRKAA1) | S486 | Uniprot |
| Q13131 (PRKAA1) | T488 | Uniprot |
| Q7Z3C6 (ATG9A) | S761 | Uniprot |
| Q86WV6 (TMEM173) | S366 | Uniprot |
| Q8N122 (RPTOR) | S792 | Uniprot |
| Q8N122 (RPTOR) | S855 | Uniprot |
| Q8N122 (RPTOR) | S859 | Uniprot |
Research Backgrounds
Serine/threonine-protein kinase involved in autophagy in response to starvation. Acts upstream of phosphatidylinositol 3-kinase PIK3C3 to regulate the formation of autophagophores, the precursors of autophagosomes. Part of regulatory feedback loops in autophagy: acts both as a downstream effector and negative regulator of mammalian target of rapamycin complex 1 (mTORC1) via interaction with RPTOR. Activated via phosphorylation by AMPK and also acts as a regulator of AMPK by mediating phosphorylation of AMPK subunits PRKAA1, PRKAB2 and PRKAG1, leading to negatively regulate AMPK activity. May phosphorylate ATG13/KIAA0652 and RPTOR; however such data need additional evidences. Plays a role early in neuronal differentiation and is required for granule cell axon formation. May also phosphorylate SESN2 and SQSTM1 to regulate autophagy.
Autophosphorylated. Phosphorylated under nutrient-rich conditions; dephosphorylated during starvation or following treatment with rapamycin. Under nutrient sufficiency, phosphorylated by MTOR/mTOR, disrupting the interaction with AMPK and preventing activation of ULK1 (By similarity). In response to nutrient limitation, phosphorylated and activated by AMPK, leading to activate autophagy.
Acetylated by KAT5/TIP60 under autophagy induction, promoting protein kinase activity.
Cytoplasm>Cytosol. Preautophagosomal structure.
Note: Under starvation conditions, is localized to puncate structures primarily representing the isolation membrane that sequesters a portion of the cytoplasm resulting in the formation of an autophagosome.
Ubiquitously expressed. Detected in the following adult tissues: skeletal muscle, heart, pancreas, brain, placenta, liver, kidney, and lung.
Interacts with GABARAP and GABARAPL2. Interacts (via C-terminus) with ATG13. Part of a complex consisting of ATG13, ATG101, ULK1 and RB1CC1. Associates with the mammalian target of rapamycin complex 1 (mTORC1) through an interaction with RPTOR; the association depends on nutrient conditions and is reduced during starvation. Interacts with FEZ1; SCOC interferes with FEZ1-binding. Interacts with TBC1D14. Interacts (phosphorylated form) with TRIM5. When phosphorylated at Ser-317, interacts with MEFV and BECN1 simultaneously. Interacts with TRIM21 and IRF3, in the presence of TRIM21. Interacts with SESN2. Interacts with SQSTM1. Interacts with C9orf72. Interacts with WDR45.
Belongs to the protein kinase superfamily. Ser/Thr protein kinase family. APG1/unc-51/ULK1 subfamily.
Research Fields
· Cellular Processes > Transport and catabolism > Autophagy - animal. (View pathway)
· Environmental Information Processing > Signal transduction > mTOR signaling pathway. (View pathway)
· Environmental Information Processing > Signal transduction > AMPK signaling pathway. (View pathway)
· Organismal Systems > Aging > Longevity regulating pathway. (View pathway)
References
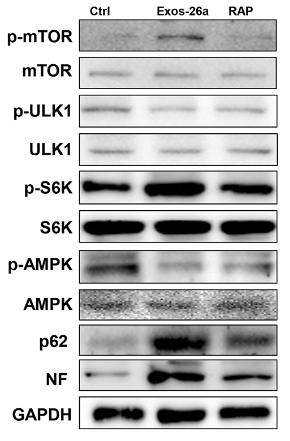
Application: WB Species: rat Sample: PC13 cells
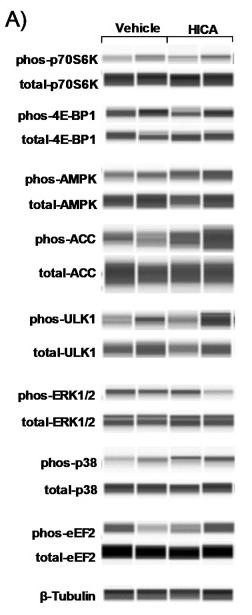
Application: WB Species: Mice Sample:
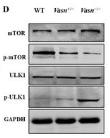
Application: WB Species: Mouse Sample: liver
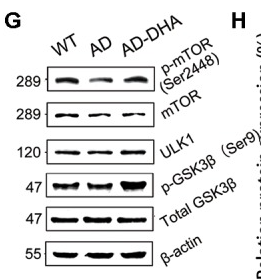
Application: WB Species: Mouse Sample: brain tissue
Restrictive clause
Affinity Biosciences tests all products strictly. Citations are provided as a resource for additional applications that have not been validated by Affinity Biosciences. Please choose the appropriate format for each application and consult Materials and Methods sections for additional details about the use of any product in these publications.
For Research Use Only.
Not for use in diagnostic or therapeutic procedures. Not for resale. Not for distribution without written consent. Affinity Biosciences will not be held responsible for patent infringement or other violations that may occur with the use of our products. Affinity Biosciences, Affinity Biosciences Logo and all other trademarks are the property of Affinity Biosciences LTD.